Abstract
The advent of small molecule-based targeted therapy has improved the treatment of both acute and chronic leukemias. Resistance to small molecule inhibitors has emerged as a common theme. The most frequent mode of acquired resistance is the acquisition of point mutations in the kinase domain. FLT3 inhibitors have improved response rates in FLT3-mutated acute myeloid leukemia (AML). The occurrence of the ATP-binding site and activation loop mutations confers varying degrees of resistance to the individual FLT3 inhibitors. Second-generation FLT3 inhibitors such as crenolanib may overcome the resistance of these mutations. Furthermore, nonmutational mechanisms of resistance such as prosurvival pathways and bone marrow signaling may be upregulated in FLT3 inhibitor-resistant AML with secondary kinase domain mutations. More recently, point mutations conferring resistance to the Bruton tyrosine kinase inhibitor ibrutinib in chronic lymphocytic leukemia, arsenic trioxide in acute promyelocytic leukemia, and the BH3-mimetic ABT199 in lymphoma have been identified. In chronic myeloid leukemia, the emergence of tyrosine kinase domain mutations has historically been the dominant mechanism of resistance. The early identification of secondary point mutations and their downstream effects along with the development of second- or third-generation inhibitors and rationally designed small molecule combinations are potential strategies to overcome mutation-mediated resistance.
Introduction
Advances in the molecular characterization of leukemia have resulted in the identification of molecular mutations with prognostic and therapeutic impact in leukemia.1,2 Perhaps the most in-depth evaluation of the impact of molecular mutations has been performed in acute myeloid leukemia (AML).3-6 A number of mutated or deregulated genes of prognostic importance have been identified.2,4 These mutations have also enhanced our ability to select optimal treatments. There are 3 predominant ways by which molecular mutations impact therapy in leukemia. First, mutated or aberrantly expressed genes are potential targets for small molecule inhibitors or monoclonal antibodies. The best example of successful targeted therapy in leukemia (and cancer therapy in general) is the use of tyrosine kinase inhibitors (TKIs) in chronic myeloid leukemia (CML) and FIP1L1-platelet-derived growth factor receptor (PDGFRα)-induced myeloproliferative diseases such as chronic eosinophilic leukemia and hypereosinophilic syndrome.7-10 More recently, small molecule inhibitors targeting FMS-like tyrosine kinase-3 (FLT3) in AML and Bruton tyrosine kinase (BTK) or the phosphatidylinositol 3-kinase kinase δ isoform (PI3Kδ) in chronic lymphocytic leukemia (CLL) have shown clinical efficacy.11-15 Second, specific molecular abnormalities stratify patients to risk-adapted strategies and aid in the selection of ideal postremission therapy (eg, allogeneic stem cell transplant for molecular poor-risk patients with AML).3 Third, the presence of mutations that regulate DNA methylation and chromatin structure may define epigenetically distinct forms of leukemia, thus potentially assisting in the identification patients with a higher likelihood of responding to epigenetic therapy.16
Molecular technologies used for the detection of mutations are generally polymerase chain reaction (PCR) based and can be broadly divided into nonsequencing technologies and sequencing technologies. Selection of the technique depends on many factors including the type of mutation and the distribution of mutation/s within the gene.17 Nonsequencing technologies that use an electrophoresis-based evaluation of specific DNA properties (eg, size, denaturation) generally involve an end point PCR and visual/analog detection and tend to be qualitative or semiquantitative in nature, with relatively low analytical sensitivity. Among these, fragment analysis by capillary gel electrophoresis is a popular method (NPM1 and FLT3 mutations).17 Incorporation of fluorescently labeled primers and probes allows real-time detection of PCR amplification in techniques such as real-time quantitative PCR [tranlocations such as t(15;17)], allele-specific PCR (for single nucleotide variants such as KIT p.D816V, JAK2 p.V617F).17,18 Real-time PCR-based techniques can achieve high levels of sensitivity (105) for minimal residual disease monitoring and detection of emerging clones. Real-time measurement of differences in the melting properties forms the basis of high-resolution melting curve analysis used in screening of relatively low-frequency mutations such as IDH1, IDH2, and DNMT3A.19 Multiplexing and multianalyte approaches in real-time PCR are gradually emerging to allow simultaneous detection of many single nucleotide variants and translocations at high analytical sensitivity.20,21 In comparison with the nonsequencing technologies listed above, which rely on physical properties of the PCR product or specificity of primer/probe sequences, the sequencing-based technologies such as pyrosequencing (best suited for short stretches of DNA such as codons 12, 13, and 61 in KRAS/NRAS; sensitivity: 5-10%), Sanger sequencing (best suited for genes with nonhotspot mutations such as CEBPA; sensitivity: 10-20%), and recent next-generation sequencing (NGS; sensitivity for clinical use: ∼5%) directly sequence the target DNA to identify changes compared with the reference sequence. Although Sanger sequencing has traditionally been considered a gold standard in sequencing, it is limited by low sensitivity, low multiplexing, and low throughput. NGS or third-generation sequencing overcomes these limitations by being able to simultaneously sequence multiple DNA fragments at a high sequencing depth. The ability to detect known and novel mutations in routine clinical testing has made this a cost-effective method of choice for diagnostic multigene profiling and has facilitated rapid adaptation of newly discovered and clinically relevant genes in routine clinical testing.22-24 Genes that were too cumbersome for routine clinical testing such as ASXL1, RUNX1, EZH2, and TET2 can be routinely sequenced on an NGS-based platform. At the same time, techniques such as droplet/digital PCR are enabling ultrasensitive detection of mutations for minimal residual disease monitoring and early detection.25,26 Overall, the advances in molecular technologies are paving a way for early detection of resistance mutations in clinically significant genes in post-therapy settings in routine clinical care.
Despite the recent advances in genomic technologies, there are hurdles to the widespread implementation of targeted therapies in clinical practice.4,27 These include discriminating driver from passenger lesions, understanding the biological impact of downstream pathways, defining the prognostic and predictive value of individual mutations in a patient with multiple molecular mutations, and failure to control proliferation of resistant clones (either preferentially selected or spontaneously mutated) on exposure to targeted agents. A common limiting factor to sustained efficacy of these molecularly targeted therapies has been the emergence of resistance in a significant proportion of the patients. A number of resistance mechanisms have been defined, including secondary mutations in the target kinase, thereby altering the conformational state and binding ability of the inhibitor, increased target ligand or receptor expression, mutations or upregulation in parallel prosurvival pathways conferring off-target resistance, and altered pharmacokinetics (CYP3A4, liver metabolism, and gut and intravenous absorption; Table 1).28-32 The most common of these involves point mutations (preexistent or acquired) in the target kinase that prevent or reduce the affinity of binding of the inhibitor to the target kinase. Secondary mutations in the FLT3 tyrosine kinase domain (TKD) in AML and Bcr-Abl kinase domain (KD) mutations in CML and acute lymphoblastic leukemia are well-known causes of resistance to TKIs.30,33-35 Recently, BTK point mutations affecting ibrutinib binding to its target (C481S) and gain-of-function mutations in a BTK-related molecule (PLCγ2) resulting in B-cell receptor (BCR) activation independent of BTK have been identified as mediators of resistance to the BTK inhibitor ibrutinib in CLL.36,37 Similarly, mutations in the promyelocytic leukemia (PML) locus (A216V, A216T, S214L, L217F, and S220G) conferring resistance to arsenic trioxide (ATO) in acute promyelocytic leukemia (APL)38,39 ; BCL-2 BH3 domain mutations (F101C and F101L) conferring resistance to the BH3 mimetic ABT-199 in lymphoma40 ; ATP-binding site mutations (T674I and D842V) conferring resistance to imatinib in chronic eosinophilic leukemia41,42 ; and Janus kinase (JAK)1 KD (Phe 958 and Pro 960) and JAK2 domain (Tyr931) mutations conferring resistance to JAK inhibitors, including ruxolitinib, have recently been described.43 In this context, it is important to note that, unlike FLT3-mutated AML or Bcr-Abl-mutated CML, BCR-mediated proliferation in CLL and BCL-2 signaling in lymphoma are not driven by a specific mutation in a target kinase but instead by upregulation of prosurvival kinase pathways such as Pi3K/AKT, mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK), and nuclear factor (NF)-κB in the lymphoid tissue. Additionally germ-line polymorphisms may be identified on sequencing. These polymorphisms, although different from mutations, may impact the propensity to respond to particular treatment regimens, including targeted therapy in leukemias.44-46 Furthermore, these polymorphisms may impact prognosis independent of therapy.
Development of resistance to these targeted agents demonstrates the plasticity of cancer cells that allows them to adapt to therapeutic pressure with the emergence of resistant clones. This ability of cancer cells to adapt is based on the heterogeneity within the malignant clone (intratumor heterogeneity), where ongoing somatic mutations promote continuous diversification, leading to genetically distinct subclones that compete for resources and are selected by their fitness to adapt to their environment and to therapeutic pressure. For example, studies in AML,47 pediatric acute lymphoblastic leukemia,48 and in CLL49 demonstrated clonal evolution with selection and expansion of ancestral leukemia subclones at the time of relapse that also acquired additional genetic lesions. In this review, we discuss the molecular and clinical consequences of mutations in specific targets, such as in target kinases or in related pathway kinases, which can interfere with drug binding or provide an alternative mechanism of pathway activation, how they impact targeted therapy in patients with leukemia, and potential strategies to overcome drug resistance related to these mechanisms with a focus on FLT3 inhibitors in AML, BTK inhibitors in CLL, TKIs in CML, and ATO in APL.
FLT3 inhibitor resistance in AML
Activating mutations in FLT3 are observed in ∼30% of adult AML patients. The 2 leading types of mutations include internal tandem duplications in the juxtamembrane domain (ITD; 17-34%) and mutations in the TKD activation loop (∼7%).50
Several small molecule FLT3 inhibitors currently undergoing evaluation in phase 1, 2, and 3 trials have shown promising activity as single agents and in combination with hypomethylating agents or chemotherapy. These include quizartinib, sorafenib, midostaurin, lestaurtinib, and crenolanib.11-13,51-55 These TKIs act as direct inhibitors of FLT3 via competitive inhibition of ATP-binding sites in the FLT3 receptor KD.56,57 The variations in conformational states (inactive vs active) of the FLT3 KDs have led to the different types of FLT3 inhibitors. Most FLT3 inhibitors including quizartinib, sorafenib, and midostaurin target the inactive conformation (type II inhibitors). However, next-generation FLT3 inhibitors, such as crenolanib, target both the inactive and active conformational states (type I inhibitor).58
FLT3 inhibitor resistance and strategies to overcome resistance
As opposed to CML, where responses to TKI therapy are rapid and durable, responses to FLT3 inhibitors are transient, lasting for 3 to 6 months (except among patients who undergo transplantation) due to the emergence of resistance.30,32 The primary cause of resistance is the acquisition of point mutations in the ATP binding region of the FLT3 KD, thereby altering the conformational state and weakening the binding affinity to specific FLT3 inhibitors. Nonmutational mechanisms of resistance include upregulation of parallel prosurvival pathways including the MEK/ERK, PI3K/Akt/mammalian target of rapamycin (mTOR), FOXO3A, and signal transducer and activator of transcription (STAT)5/PIM pathways, upregulation of the FLT3 ligand or FLT3 receptor, mutations in other kinases (eg, CBL), activation of antiapoptotic proteins BCL2, MCL1, and BCL-x(L), and tumor microenvironment/stroma-mediated resistance30,32,59 (Figure 1A-B). These mechanisms of resistance and ongoing strategies to overcome resistance to FLT3 inhibitors will be discussed.
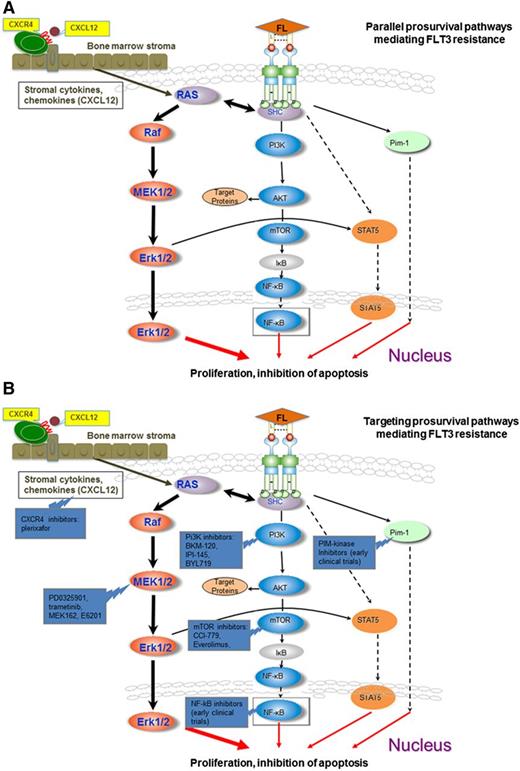
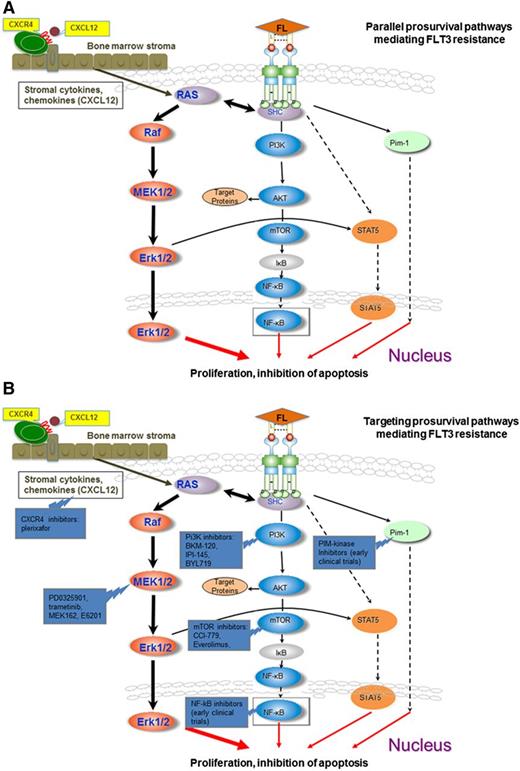
Activation and targeting of prosurvival pathways mediating FLT3 resistance. (A) The downstream effectors pERK, pAKt, pS6K, and pSTAT are not sufficiently inhibited despite FLT3 signaling blockade in sorafenib-resistant FLT3 cell lines harboring acquired point mutations in the tyrosine kinase domains. The MEK/ERK pathway likely plays a central role in this feedback modulation with smaller contributions from the PI3K/AKT and STAT5 pathways. Circulating blasts are dependent on FLT3 signaling for activation of the MEK/ERK pathways. Stromal cytokines may activate the MEK/ERK pathway in stromal leukemia blasts independent of FLT3-ITD signaling, such as through CXCL12/CXCR4 interactions, conferring resistance to the clinically available FLT3 inhibitors. (B) Concomitant inhibition of CXCR4 (plerixafor), Pi3K (Pi3K inhibitors: BKM-120, IPI-145, BYL-719), MEK/ERK (PD0325901, trametinib, MEK-162, E6201), AKT/mTOR (mTOR inhibitors: CCI-779, everolimus), NF-κB inhibitors (early clinical trials), PiM-kinase inhibitors (early clinical trials), and FLT3 produces a synergistic effect in sorafenib-resistant cell lines.
Activation and targeting of prosurvival pathways mediating FLT3 resistance. (A) The downstream effectors pERK, pAKt, pS6K, and pSTAT are not sufficiently inhibited despite FLT3 signaling blockade in sorafenib-resistant FLT3 cell lines harboring acquired point mutations in the tyrosine kinase domains. The MEK/ERK pathway likely plays a central role in this feedback modulation with smaller contributions from the PI3K/AKT and STAT5 pathways. Circulating blasts are dependent on FLT3 signaling for activation of the MEK/ERK pathways. Stromal cytokines may activate the MEK/ERK pathway in stromal leukemia blasts independent of FLT3-ITD signaling, such as through CXCL12/CXCR4 interactions, conferring resistance to the clinically available FLT3 inhibitors. (B) Concomitant inhibition of CXCR4 (plerixafor), Pi3K (Pi3K inhibitors: BKM-120, IPI-145, BYL-719), MEK/ERK (PD0325901, trametinib, MEK-162, E6201), AKT/mTOR (mTOR inhibitors: CCI-779, everolimus), NF-κB inhibitors (early clinical trials), PiM-kinase inhibitors (early clinical trials), and FLT3 produces a synergistic effect in sorafenib-resistant cell lines.
Mutational resistance to FLT3 inhibitors
Cools et al60 and Heidel et al61 initially identified point mutations in the FLT3-ITD KD as a mediator of resistance to the FLT3 inhibitor midostaurin. In these studies, the cells with mutations in the KD had similar levels of kinase activity to the FLT3-ITD cells and comparable levels of membrane and total protein expression of receptors.61 However, mutations in the KD hampered the ability of midostaurin to adequately bind and inhibit the kinase activity either by creating a steric clash with the inhibitor resulting in decreased binding affinity, removal of favorable hydrophobic contacts, or destabilization of the conformation of the hinge segment or P-loop, which makes contact with the midostaurin.60 Smith et al62 used subcloning and sequencing to study paired pretreatment and relapse samples in 8 patients who initially responded but subsequently relapsed with quizartinib therapy. Analysis of individual FLT3-ITD alleles revealed mutations at the time of relapse that were not detected before treatment in all 8 cases. This suggests that resistance via secondary mutations in the FLT3 is due to novel mutations occurring during FLT3 inhibitor therapy rather than selective expansion of a preexisting clone under the pressure of FLT3 inhibitor therapy. Homology modeling has identified 2 main types of FLT3 mutations, namely TKD1 mutations involving the ATP binding (hinge regions) and the TKD2 mutations involving the activation loop.63
Zhang et al64 have shown that a mutation in either TKD1 or TKD2 was sufficient to provide resistance to sorafenib-induced apoptosis and to upregulate phosphorylated FLT3. The TKD2 mutations conferred a greater degree of resistance than analogous TKD1 mutations. However, compound mutations in both TKD1 and TKD2 displayed the highest resistance. Furthermore, although sorafenib retained some degree of effectiveness in suppression of the phosphorylated FLT3 in TKD mutant cell lines, this was not associated with parallel suppression of the downstream prosurvival pathways. For example, elevated levels of phosphorylated extracellular signal-related kinase (pERK) were noted in TKD2 mutant cell lines treated with sorafenib, and persistent levels of pAKT, pS6K, and pSTAT5 were noted in compound mutant cell lines, suggesting feedback upmodulation of MAPK/ERK and AKT/S6K signaling pathways. The TKD mutations also conferred resistance to other type 2 FLT3 inhibitors such as quizartinib but retained sensitivity to type 1 TKIs such as midostaurin and crenolanib, suggesting that mutations in TKD2 directly interfere with the binding of type 2 TKIs, whereas retaining sensitivity to type 1 TKIs. In fact, extremely low concentrations of crenolanib markedly suppressed phosphorylation of FLT3 and its downstream effectors ERK and AKT in the sorafnib-resistant cell lines. In turn, the combination of type 1 (sorafenib) and type 2 (crenolanib) TKIs caused profound synergistic apoptosis in sorafenib-resistant cells. Similar synergism was reproducible by combining other type 1 and type 2 TKIs (eg, midostaurin and sorafenib) or combining 2 type 1 TKIs (eg, crenolanib and midostaurin) but not by combining 2 type 2 TKIs.
The resistance profiles that emerge with FLT3 inhibitors are nonoverlapping.33 Individual FLT3 kinase inhibitors have distinct inhibitory potencies against different activating TKD mutations.60,65 Cools et al identified TKD1 mutations N676D, F691I, and G697R in a screen for resistance mutations using midostaurin.60 Kancha et al demonstrated that type 2 FLT3 inhibitors (sunitinib and sorafenib) were able to overcome midostaurin-resistant mutations at therapeutic levels.66 Thus, the type 2 FLT3 inhibitors (sorafenib and sunitinib) may be able to overcome TKD1 mutations that confer resistance to type 1 FLT3 inhibitors (midostaurin). Altogether, these findings suggest that resistance to 1 type FLT3 inhibitor may be overcome by an alternate type FLT3 inhibitor or by combining FLT3 inhibitors. Thus, rational combinations of FLT3 inhibitors or switching FLT3 inhibitors may overcome mutational resistance, and these approaches are currently being evaluated in clinical trials.
In vitro assays suggest that quizartinib may be the most potent FLT3 inhibitor currently in phase 2/3 studies. Among 76 patients with relapsed/refractory AML treated with quizartinib 12 to 450 mg/day, 30% achieved marrow responses, lasting for a median of 3.5 months.13 However, mutations conferring to quizartinib have been identified. Smith et al performed a previously validated in vitro saturation mutagenesis67 assay to identify quizartinib resistance conferring mutations at 4 residues in the kinase domain of FLT3-ITD (F691, D835, Y842, and E608). Mutations at 3 of these residues: gatekeeper residue (F691I) and activation loop residues (D835, Y842) conferred a high degree of resistance to quizartinib in proliferation and cell-based biochemical assays.67 Subcloning and sequencing68 in paired pretreatment and relapsed samples showed the emergence of drug-resistant point mutations in 8 FLT3-ITD mutated patients who initially achieved response but subsequently relapsed on quizartinib therapy.62 In all 8 cases, the mutations identified at the time of relapse were not detected before treatment. Mutations identified by sequencing were confined to the D835 and F691 residues and included D835Y in 3 cases, D835V in 2 cases, and F691L in 3 cases. One of the patients acquired polyclonal mutations with both F691L and D835V mutations detected on separate FLT3-ITD sequences. A novel D835F mutation conferring a high degree of resistance to both quizartinib and sorafenib was detected. D835 was not identified on the saturation mutagenesis because its creation requires a 2-nucleotide substitution. To further characterize resistance-conferring mutations, they performed focused interrogation of FLT3-ITD alleles using third-generation sequencing with single-molecule real-time sequencing.69 This assay produced a large number of reads (range, 19-930) of the ITD region and KD with an average read length of >1 kb. Resistance-conferring mutations were confirmed in all 8 patients with the frequency of individual codon substitutions within FLT3-ITD ranging from as low as 3% to 51%. Interestingly, polyclonal mutations were identified in 4 of 8 patients compared with 1 of 8 using standard sequencing. These studies demonstrate that improved sequencing techniques allow the identification of novel mutations or emergence of polyclonal mutations that were missed by less sensitive technologies such as saturation mutagenesis and second-generation sequencing, thereby revealing the true genetic complexity of drug-resistant disease and the immense selective pressure exerted by the FLT3 inhibitors.
Limitations in clinically available FLT3 inhibitors have led to the development of novel FLT3 inhibitors and FLT3 inhibitor combinations. Crenolanib possesses activity against both TKD1 and TKD2 mutants, including the D835H and D835Y mutants,55,58,70 and is in clinical trials.71,72 The F691L “gatekeeper” mutation causes resistance to quizartinib by disrupting the interaction between the aromatic side chain of the phenylanaline residue and the benzo-imidazol-thiazol moiety of quizartinib. Ponatinib is structurally different from quizartinib and does not depend on this interaction. Ponatinib has shown preclinical activity and responses in a phase 1 study in quizartinib-resistant patients harboring the gatekeeper F691I mutation.73
Activation of parallel prosurvival MEK/ERK and AKT/S6K pathways
Zhang et al64 has shown that the inhibition of phosphorylated FLT3 was not sufficient to inhibit its downstream effectors pERK, pAKT, pS6K, and pSTAT in sorfenib-resistant cell lines harboring acquired point mutations in the TKDs of the FLT3 gene. This suggests the role of feedback upmodulation via the mitogen-activated protein kinase kinase (MEK)/ERK and/or AKT/S6K pathways in sorafenib-resistant cell lines (Figure 1A). In turn, concomitant MAPK blockade with a MEK inhibitor (CI-1040), or of AKT/mTOR pathway with mTOR antagonist (CCI-779), induced a synergistic proapoptotic effect in sorafenib-resistant cell lines (Figure 1B). The combination with a MEK inhibitor produced a more synergistic effect than with the mTOR antagonist. Maximum synergism was demonstrated by concomitant blockade of FLT3, MEK, and AKT pathways. Similar findings have been reported by Yang et al,74 who noted upregulation of the ERK pathway in FLT3 mutant cell lines cocultured with stroma and/or FLT3 ligand despite FLT3 inhibition. They noted that FLT3 mutant cells cocultured with either bone marrow stroma or exogenous FLT3 ligand demonstrated pronounced ERK phosphorylation. The ERK phosphorylation could not be inhibited, even at doses of quizartinib or sorafenib that fully inhibited phosphorylation of AKT and FLT3. MEK inhibitors (PD0325901 and trametinib) induced minimal apoptosis on their own, but when combined with quizartinib or sorafenib, the results were synergistic. These findings suggest that concomitant inhibition of FLT3 and ERK activation might abrogate stromal protection and induce apoptosis of leukemic cells in the stromal niche. E6201 is a dual FLT3/MEK inhibitor with potent activity against TKD1, TKD2, and compound mutations.75 A clinical trial with E6201 (a dual FLT3/MEK inhibitor) is planned to test this hypothesis.
Bone marrow microenvironment-mediated resistance
FLT3 inhibitors seem to have a more profound impact on peripheral blasts compared with bone marrow blasts.32 A potential explanation for the reduced bone marrow response may be microenvironmental resistance to FLT3 inhibitors, in part via protection afforded by CXCL12 (stromal-derived factor 1 [SDF-1])/CXCR4 signaling.76-78 It is postulated that disrupting the CXCL12/CXCR4 axis with a highly potent CXCR4 antagonist will disrupt the interaction of the malignant blasts with the bone marrow microenvironment and may augment the antileukemic effect of chemotherapy and kinase inhibitors including FLT3-ITD inhibitors. Zeng et al59 have shown that CXCR4 inhibition enhances the sensitivity of FLT3-mutated leukemic cells to the apoptogenic effects of the FLT3 inhibitor sorafenib. CXCR4 expression was significantly higher in FLT-ITD AML than in FLT3 wild-type AML. Furthermore, Rombouts et al77 noted that the prognostic significance of FLT3-ITD with respect to relapse-free survival was lost when CXCR4 expression in the CD34+ subset of leukemic cells is added to the multivariate analysis, suggesting that the poor prognosis of FLT3-ITD AML might be subordinate to the increased CXCR-4 expression. The combination of FLT3 inhibitor sorafenib CXCR4 antagonist AMD3100 demonstrated an impressive overall response rate >70% in heavily pretreated AML patients with FLT3-ITD mutations, with several long-lasting responses even in patients previously treated with FLT3 inhibitors.59,79
BTK inhibition in CLL and other lymphoid malignancies
In normal B cells, BCR signaling plays a crucial role for expansion and maturation of antigen-specific B cells. Similarly, in several B-cell malignancies, such as CLL and diffuse large cell B-cell lymphoma (DLBCL), BCR signaling also plays a central role in the pathogenesis, even though the mechanisms of BCR stimulation are heterogeneous and to some degree controversial.80-83 Two different mechanism of BCR pathway activation in B-cell lymphomas are recognized: antigen-dependent and -independent activation. For example, there can be activating mutations in the BCR pathways, such as mutations in the coiled-coil (CC) domain of CARD11 in DLBCL or activating mutations of the immunoreceptor tyrosine-based activation motifs within the CD79B and CD79A signaling modules of the BCR, as in activated B-cell DLBCL.82,84 In CLL, on the other hand, BCR pathway activation is viewed as antigen dependent, resulting from BCR ligation via antigens (autoantigens and/or microbial antigens) that are present in the microenvironment. Furthermore, unlike FLT3-mutated AML or Bcr-Abl-mutated CML, BCR signaling in CLL is not driven by a specific mutation but instead by upregulation of prosurvival pathways such as Pi3K/AKT, MAPK/ERK, and NF-κB in the lymph nodes and bone marrow.
BTK is a nonreceptor tyrosine kinase of the Tec kinase family that plays a central role in BCR signaling. On BCR activation, BTK becomes activated by other tyrosine kinases, such as Lyn and SYK, resulting in activation of transcription factors necessary for B-cell proliferation and differentiation.85 In addition to its role in BCR signaling, BTK is involved in signaling of other receptors related to B-cell migration and adhesion, such as chemokine receptors (CXCR4 and CXCR5) and adhesion molecules (integrins).86-88 Ibrutinib, previously called PCI-32765, binds irreversibly to a cysteine residue (Cys-481) in the BTK kinase domain and inhibits BTK phosphorylation and its enzymatic activity.89 Byrd et al14 recently reported that single agent ibrutinib induces an overall response rate of 71% in CLL patients with relapsed or refractory disease and an additional 15% to 20% of patients had a partial response with lymphocytosis. The response was independent of clinical and genomic risk factors present before treatment. At 26 months, the estimated progression-free survival rate was 75% and the rate of overall survival was 83%. Wang et al reported single agent efficacy of ibrutinib in patients with relapsed or refractory mantle cell lymphoma, with a complete response rate of 21% and a partial response rate of 47%.90
Ibrutinib resistance
Development of resistance to ibrutinib is closely correlated with high-risk disease. Especially patients with del(17p) or TP53 mutations are at increased risk for developing resistance, with an approximate rate of 13% of patients becoming resistant at a median follow-up of 26 months.14 Among these, patients with Richter’s transformation were more common than patients with CLL progression. Two reports recently examined mechanism of resistance to ibrutinib in CLL patients with CLL progression and described point mutations in BTK (C481S)36,37 that disrupt ibrutinib binding and in its related pathway member PLCG2 (R665W, L845F, S707Y) that can activate the BCR pathway independently from BTK as mechanisms of ibrutinib resistance. It is unclear whether the expansion of these resistant clones is due to preexisting subclones that are selected by the continuous therapy with ibrutinib or rather represent de novo mutations that occurred during ibrutinib therapy. Based on computational evolutionary models, analyzing measured kinetic parameters of patients, it was predicted that BTKi-resistant mutants exist before initiation of ibrutinib therapy, although they only comprise a miniscule portion of the overall disease burden. Depending on growth and death rates of the individual CLL cell clone, the time to disease progression may become predictable using such models.91 Development of ibrutinib resistance and risk factors for developing resistance has important clinical consequences. If resistance is likely to develop in a high-risk CLL patient responding to ibrutinib, allogeneic transplantation should be considered while still responding to ibrutinib92 rather than delaying the stem cell transplantation, given the generally more aggressive behaviors and poor prognosis once ibrutinib resistance has occurred. Whether del(17p) or TP53 mutations alone,93 or rather additional complex cytogenetic abnormalities are more appropriate for predicting the risk for ibrutinib resistance, is at this time controversial and hopefully will be answered by prospective studies.
Overcoming ibrutinib-resistant mutations in lymphoid malignancies
Currently, there is no established regimen for treating CLL patients who become ibrutinib resistant. If feasible, allogeneic stem cell transplantation should be considered in patients who respond to the next line of therapy. Different second-generation BTK inhibitors or other kinase inhibitors targeting other kinases within the BCR signaling pathway are currently being explored. IPI-145, an inhibitor of PI3K p110δ and p110γ, antagonizes prosurvival signaling in BCR-activated CLL cells and promotes apoptosis in CLL cells while sparing normal B cells.94 IPI-145 completely inhibited PI3K signaling and downstream AKT phosphorylation even in patients harboring C418S mutations that confer resistance to ibrutinib.95 IPI-145 is currently being evaluated for therapy of CLL. Alternatively, treatment with the BCL-2 antagonist ABT-199 will be explored, and in the absence of clinical trial options, some patients have been treated with combinations of anti-CD20 mAbs and high-dose corticosteroids. Hopefully, detailed genetic and molecular analyses of BTK-resistant patients and signals from ongoing clinical trials with new agents will lead to a more rational treatment approach.
TKI resistance in CML
The best example of effective targeted therapy in cancer is the use of TKIs that target the Bcr-Abl1 oncoprotein and inhibit its kinase activity. Imatinib mesylate, a potent inhibitor of the tyrosine kinases ABL, ARG, PDGFR, and cKIT, targets the Bcr-Abl1 oncoprotein and was the first TKI to receive approval for the treatment of patients with CML in chronic phase (CML-CP).7,96
Imatinib-resistant Bcr-Abl KD mutations
Incidence of imatinib-resistant Bcr-Abl KD mutations
Although the results with imatinib are impressive, only 55% of patients remained on therapy at the 8-year follow-up time.97 The 2 main reasons for discontinuing imatinib-based therapies were resistance and intolerance/noncompliance. A frequent mechanism of resistance to imatinib is the occurrence of point mutations in the Bcr-Abl KD, although Bcr-Abl amplification and impaired signaling pathways may contribute to the resistance.98 Other possible Bcr-Abl independent mechanisms that incite resistance include reduced bioavailability of imatinib within Ph-positive cells, genomic amplification of Bcr-Abl, clonal cytogenetic changes, loss of functional p53, and activation of alternative signaling pathways that promote cell survival and proliferation.99,100 Preexisting and evolving Bcr-Abl mutations could be identified at diagnosis (pre-TKI) using sensitive sequencing techniques,101,102 However, pre-TKI KD mutations appear to be more prevalent in Sokal high-risk patients and accelerated/blast phase CML.103,104 For example, KD mutations prior to initiation of TKI were identifiable by direct sequencing in 8 of 13 (62%) Sokal high-risk, 1 of 6 Sokal intermediate-risk, and 0 of 24 Sokal low-risk patients.102 Willis et al104 did not detect Bcr-Abl KD mutations in unselected, imatinib naïve CML-CP patients; however, ∼30% of accelerated phase (CML-AP) and 25% of blast phase CML (CML-BP) imatinib-naïve patients harbored mutations. Therefore, Bcr-Abl KD mutation analysis in newly diagnosed CML-CP may be reserved for Sokal high-risk or accelerated/blast phase patients.
Routine monitoring of all patients on imatinib therapy is not recommended. Lahaye et al105 evaluated long-term outcome, rates of response, and resistance in 300 patients with Bcr-Abl-positive leukemia. They identified Bcr-Abl mutations in 45% of patients (including CML-CP, CML-AP, and CML-BP) with hematologic resistance to imatinib.105 The frequency of KD mutations among front-line imatinib resistance patients differs in different phases of CML, ranging from 25% to 30% in CML-CP to 70% to 80% in CML-BP.106,107 The EuropeanLEukemia Net defines suboptimal response as the failure to achieve specific response milestones at specific time points or by the loss of a previously achieved response milestone. The incidence of KD mutations in suboptimal responders is 15%, with the highest incidence (24%) occurring among patients who fail to achieve a major cytogenetic response by 12 months.108
Impact of imatinib-resistant BCR-ABL KD mutations
More than 100 different BCR-ABL KD mutations have been identified. However, 15 mutations comprise 85% of all those detected (supplemental Figure 1, available on the Blood Web site).34 A mutant clone does not necessarily have a proliferative advantage nor does it necessarily confer resistance in CML.109 Only a subset of mutations (G250E, Y253H, E255K/V, V299L, T315I, F317L/I, F359V/I/C, H396R, E450G/V, and E459K) are associated with resistance to TKIs. The presence of a BCR-ABL KD mutation in a patient with CML on imatinib with failure, suboptimal response, or rising BCR-ABL transcript levels is an indication for change in therapy. Mildly resistant mutations (eg, M244V, M351T, or F359V) might be overcome by increasing the imatinib dose to 600 or 800 mg daily.110 Highly resistant mutants of the KD (eg, Y253F/H, E255K/V, T315I, or H396P/R) need alternative therapy with a second- or third-generation TKI, stem cell transplant, or omacetaxine. The National Comprehensive Cancer Network published guidelines for selection of treatment options based on specific KD mutations (supplemental Table 1).
Second- and third-generation TKI-resistant BCR-ABL KD mutations
Second-generation TKIs are more potent than imatinib and overcome many of the imatinib-resistant mutations. Specific mutations exhibit different levels of sensitivity to TKIs. The type of the mutation serves as a guide to the rational selection of second-generation TKIs after imatinib failure.35,111,112 Dasatinib and nilotinib have been approved for first- and second-line treatment of CML in the United States based on favorable results from the phase 3 dasatinib vs imatinib in newly diagnosed chronic myeloid leukemia in chronic-phase trial (DASISION) and evaluating nilotinib efficacy and safety in clinical trials–newly diagnosed patients (ENEST-nd) studies, respectively.8,9,113,114 Bosutinib and ponatinib are approved for second- or subsequent-line treatment of CML in the United States.115,116
Mutations rendering the leukemia resistant to second- and third-generation TKIs have emerged. Mutations Q252H, E255V, and F317L confer resistance to dasatinib; Y253H, E255K/V, and F359V confer resistance to nilotinib; G250E and E255K confer resistance to bosutinib; and E255V and H396R confer resistance to ponatinib. The T315I clone confers resistance to 4 TKIs, the exception being ponatinib. In the study by Soverini et al,117 patients who already harbor imatinib-resistant mutations had a higher incidence of relapse associated with development of further mutations on treatment with second-line (56%) or third-line TKIs (65%) compared with patients who did not harbor mutations. Cortes et al118 evaluated mutational status in 112 patients with CML who received a second-generation TKI (nilotinib or dasatinib) after imatinib failure.118 Response rates were similar among patients with and without mutations, regardless of the mutation site, except for T315I. As opposed to the study by Soverini et al, only 26% of the patients developed a new BCR-ABL KD mutation after therapy with a second or third TKI. Interestingly, they did not see a marked increase in the development of T315I mutation.
Overcoming resistance to KD mutations in CML
The KD mutations that are responsible for resistance occur at strategic locations, inhibiting TKI binding while allowing ATP binding and kinase activity.119 Because each individual TKI has specific binding characteristics, resistance profiles are specific to each TKI. The mutations may directly involve the kinase active site, thereby sterically preventing TKI binding. In other instances, the mutations may involve residues that alter the overall 3-dimensional structure of the kinase. In the case of imatinib and nilotinib, which bind BCR-ABL in its inactive conformation, mutations in residues that prevent the adoption of the inactive conformation would result in resistance to these agents.120 The second-generation inhibitors are effective against most imatinib-resistant KD mutations. Therefore, adequate mutational screening and switch to an alternate second-generation TKI with known sensitivity to the identified mutation have abrogated the impact of mutational resistance (supplemental Table 1). The only caveat to this approach is the emergence of the T315I mutation. The T315I mutation does not disturb the overall structure of the BCR-ABL protein; it affects the topology of the ATP binding region. First- and second-generation TKIs are unable to inhibit T315I mutant tyrosine kinase due to loss of the threonine residue (and its associated hydroxyl group), which are critical to binding and potency of first- and second-generation TKIs. Furthermore, introduction of the bulkier isoleucine blocks entrance of the TKIs into the hydrophobic pocket. The introduction of the third-generation TKI ponatinib has provided a viable solution to this problem. Ponantinib was specifically designed to overcome these limitations and not only inhibits BCR-ABLT315I but also inhibits native BCR-ABL and other commonly seen BCR-ABL mutations.116 Zabriskie et al121 recently demonstrated that acquired T315I-inclusive compound mutants and the I315M single mutant confer high-level resistance to TKIs, including ponatinib due to impaired drug binding. Sequential TKI treatment may result in the selection of BCR-ABL1 compound mutants, defined as a BCR-ABL1 allele harboring ≥2 mutations122 that have the potential to confer resistance to multiple TKIs and may emerge as a clinical problem in patients treated with ponatinib.
ATO in APL
ATO binds directly to cysteine residues in zinc fingers located within the B2 domain of PML protein-RARα and PML.123 The all trans-retinoic acid (ATRA) and ATO combination is safe, synergistic, and is associated with a complete response rates of 95%, a shorter time to complete response, a lower relapse rate, and a longer survival than with standard APL therapy.124
The direct binding of ATO to the PML protein is required to produce a response in APL. Mutations in the PML domain conferring resistance to ATO have been identified in the C212 and C213 domains in ex vivo models.125 Zhu et al38 identified mutations in 9 of 13 patients with arsenic-resistant disease. The mutations occurred in the PML locus (A216V, A216T, S214L, L217F, S220G), the RARα locus, or both. Isolated RARα mutations could be overcome by combination therapy with ATO and ATRA. However, patients with mutations in both RARα and PML remained refractory to the combinatorial approach. Interestingly, no mutations were identified among 22 patients with APL who were sensitive to ATO. The PML mutations seem to confer significant resistance and a poor outcome with deaths in 8 of 9 patients with PML mutations.38 Lehmann-Che et al39 recently reported the occurrence of the A216V mutation in a single patient with highly refractory APL.
Summary
The widespread application of whole exome and whole genome sequencing will result in the identification of numerous hitherto unknown molecular targets. The achievement of efficacious and durable responses with small molecule inhibitors rests on our ability to overcome the hurdle of secondary resistance. Knowledge from the CML experience is the most detailed in this regard, with established guidelines for accurate detection, quantification, and intervention for secondary KD mutations. The increasing use of small molecule inhibitors in other leukemias (AML, CLL, and APL) warrants the establishment of similar individualized diagnostic and therapeutic guidelines with intent to overcome secondary drug resistance in these disorders.
The online version of this article contains a data supplement.
Acknowledgments
This study was supported in part by MD Anderson Cancer Center Leukemia Support Grant CA016672, and generous philanthropic contributions to the MD Anderson Moon Shots Program.
Authorship
Contribution: N.D., J.C., F.R., J.A.B., M.K., K.P.P., and H.K. wrote the paper; N.D. and H.K. conceptualized and designed the review; and all authors participated in the discussion and reviewed and approved the current version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Naval Daver, The University of Texas MD Anderson Cancer Center, Department of Leukemia, 1515 Holcombe Blvd, Unit 428, Houston, TX 77030; e-mail: ndaver@mdanderson.org.