Key Points
tmTNF-α expressed on LSC and leukemia cells correlates with poor risk stratification and adverse clinical parameters.
Targeting tmTNF-α by monoclonal antibody eradicates LSC and blasts, preventing leukemia regeneration in secondary transplant in NOD-SCID mice.
Abstract
To design an effective antibody therapy to improve clinical outcomes in leukemia, the identification of novel cell surface antigens is needed. Herein, we demonstrate a role for transmembrane tumor necrosis factor-α (tmTNF-α) in leukemia. To characterize tmTNF-α expression in acute leukemia (AL), normal hematopoietic cells, and nonhematopoietic tissues, we used a monoclonal antibody, termed C1, which specifically recognizes the tmTNF-α domain. We found that tmTNF-α was preferentially expressed by AL and leukemia stem cells (LSCs). More abundant expression correlated with poor risk stratification, extramedullary infiltration, and adverse clinical parameters. Moreover, knockdown of tmTNF-α+ expression rendered leukemia cells more sensitive to chemotherapy in vitro and delayed regeneration of leukemia in NOD-SCID mice. Targeting tmTNF-α by C1 resulted in leukemia cell killing via antibody-dependent cell-mediated and complement-dependent cytotoxicity in vitro and inhibited leukemia cell growth in vivo while simultaneously sparing normal hematopoietic cells. Notably, C1 administration impaired the regeneration of leukemia in secondary serial transplantation into NOD-SCID mice. In conclusion, tmTNF-α has a favorable AL- and LSC-associated expression profile and is important for the survival and proliferation of these cells. C1-mediated targeting shows potent anti-LSC activity, indicating that tmTNF-α represents a novel target antigen in AL.
Introduction
To design effective antibody therapies for improving the clinical outcomes of leukemia patients, the identification of novel cell surface antigens is essential. There are 2 bioactive forms of tumor necrosis factor (TNF) α protein: the transmembrane form (tmTNF-α) and the secretory form (sTNF-α). The latter is produced from tmTNF-α via proteolytic cleavage by the metalloprotease TNF-α-converting enzyme.1,2 A growing body of evidence has highlighted an essential role for TNF-α in the maintenance of leukemia hematopoiesis. For example, sTNF-α signaling has been shown to promote cellular leukemia-initiating capacity in an autocrine manner.3-7 Autocrine TNF-α production supports the survival and proliferation of chronic myeloid leukemia stem and progenitor cells.8 These data strongly imply that targeting TNF-α in leukemia might represent an attractive therapeutic strategy.
Clinical trials with TNF antagonists that target TNF-α have shown promising clinical activity against certain types of solid tumors.9-13 However, no data are yet available regarding the clinical efficacy of TNF-α antagonists in hematologic malignancies. Furthermore, relatively little is known about tmTNF-α in leukemogenesis because tmTNF-α expression has been assumed to be positively correlated with sTNF-α serum levels. The expression pattern of tmTNF-α in acute leukemia (AL) and its clinical relevance are therefore of clinical interest and importance for the design of novel antibody therapies.
Previously, we successfully generated a monoclonal antibody (mAb) termed C1 that specifically recognized the tmTNF-α domain but did not bind to sTNF-α. We also showed that C1 has therapeutic activity in mouse models of breast cancer.14 In the present study, we use this antibody to determine the expression profile of tmTNF-α in AL. Importantly, our findings emphasize the clinical relevance of tmTNF-α in AL and highlight the potential antileukemia activity of C1 via targeting the tmTNF-α domain.
Materials and methods
Clinical samples and cell lines
The bone marrow (BM), BM core biopsy, and blood specimens were collected in Tongji Hospital before treatment from patients aged >14 years who had been diagnosed with acute myeloid leukemia (AML; n = 69), B or T acute lymphoblastic leukemia (B- or T-ALL; n = 30, all the cases are ALL-L2), or nonmalignant anemia (n = 30), based on the published 2008 criteria of the World Health Organization and FAB classification. The leukemia cell lines 7051-M5, 7052-M6, and le93-M3 were initially derived from patients with primary AL who remained stably transplantable in NOD-SCID mice.15 These cells and CD34+-enriched mononuclear cells harvested from healthy donors for stem cell transplantation were used for the NOD-SCID mice study. The appropriate informed consent was obtained from all the donors before specimen collection, in accordance with the Declaration of Helsinki, and all research involving human subjects was approved by the ethics committees of Tongji Hospital.
HL60 (acute promyelocytic leukemia), THP-1 (AML-M5, MLL-AF9), OCI-AML-3, Molt4 (T-ALL), and Jurkat (T-ALL) from American Type Culture Collection (Manassas, VA) and SKM-1 (myelodysplastic syndrome) from Japanese Collection of Research Bioresources Cell Bank (Ibaraki City, Osaka, Japan) were cultured in RPMI 1640, Iscove modified Dulbecco medium, or α minimum essential medium (Thermo Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (GIBCO).
NOD-SCID mice studies
All experiments with NOD-SCID mice were performed following a protocol approved by the Institutional Review Board for Animal Welfare of Tongji Hospital, and carried out at Crown BioScience Inc. (Beijing, China). The mice were intraperitoneally administered 200 μg of anti-mouse CD122 mAb (TM-β1, Rat IgG2b; BioXCell, West Lebanon, NH) 1 day before inoculation with AML cells, whereas mice inoculated with CD34+ human hematopoietic cells, obtained from normal BM (NBM), received an additional conditioning, 1.5 Gy of total body irradiation 1 day before administration of CD122 mAb. C1 or immunoglobulin (Ig) G1 (10 μg/g body weight) was intraperitoneally given twice a week, 24 hours posttransplantation. The antibody treatment lasted 6 weeks. For the survival study, the mice were treated with antibodies until they died. For homing analysis, the mice were injected with C1 or IgG1 immediately after inoculation with 7052 cells or normal CD34+ cells and euthanized 16 hours after transplantation.16,17 For the second transplantation, CD45+ leukemia cells from spleens of mice treated with C1 or IgG1 after first transplantation were inoculated into the new recipient mice. To determine the effect of C1 on NBM, the recipient mice were inoculated with CD34+ human hematopoietic cells and treated with antibodies, twice a week, for 6 weeks.
Microarray analysis
Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA) from SKM-1 stably transfected with control or tmTNF-α short hairpin RNA (shRNA), tmTNF-α+ or tmTNF-α− cells sorted from a primary AML sample, and CD45+ leukemia cells from mice treated with C1 or IgG1 after first transplantation of 7052. These were then subjected to the GeneChip probe arrays (HG-U133 Plus 2.0; Affymetrix, San Diego, CA). The Affymetrix GeneChip Command Console computer software was used to process data from the GeneChips. The signal value ratio was calculated to analyze the differentially expressed genes that were used for pathway enrichment analysis (Kyoto Encyclopedia of Genes and Genomes) with Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources (https://david.ncifcrf.gov)18 and to produce the heatmap with Cluster 3.0 and Java TreeView. The expression of typical genes selected from the microarray data were further validated by quantitative real-time polymerase chain reaction (qPCR) and western blot. Microarray data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE71459.
Statistical analysis
Data were presented as the means ± standard errors of the mean (SEMs). The differences were analyzed by the Student t test between 2 groups or 1-way analysis of variance test for >2 groups. Survival curves were compared using the log-rank test. All the P values are 2-sided. For pathway enrichment analysis with DAVID, the significance was determined using a Q value <0.25.
Results
Preferential expression of tmTNF-α in leukemia cells correlates with adverse clinical features of AL
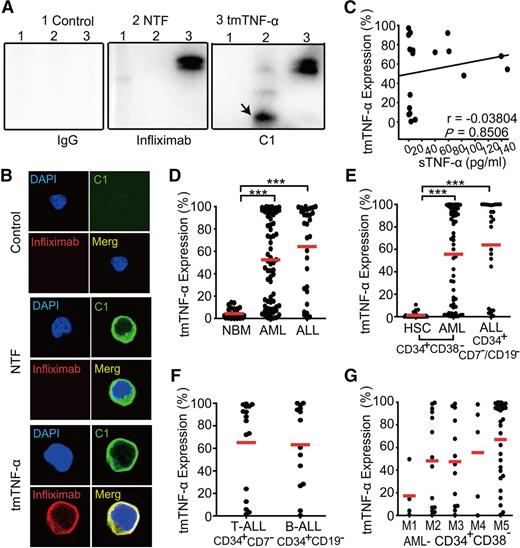
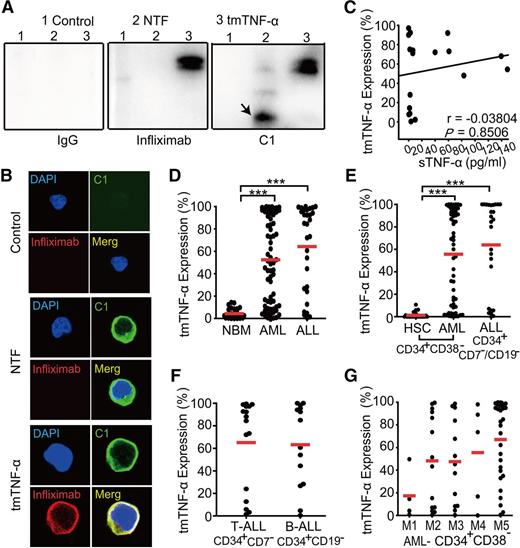
The mAb C1, which can recognize the tmTNF-α domain, but not sTNF-α, was illustrated in supplemental Figure 1A (see the Blood Web site). Our data indicate that infliximab, which was used in the clinic to target sTNF-α, could detect both recombinant sTNF-α and tmTNF-α, whereas C1 selectively recognized tmTNF-α alone (supplemental Figure 1B). Validation experiments showed that the tmTNF-α expression levels in leukemia cell lines detected by western blotting were in good agreement with those determined by flow cytometry using C1 mAb (supplemental Figure 1C-E). To further confirm the specificity of C1 antibody, tmTNF-α− Jurkat cells were stably transfected with tmTNF-α or N-terminal truncated fragment (NTF) expressing plasmid (supplemental Figure 1F). NTF is the truncated tmTNF-α molecule that does not contain the sTNF-α portion.19 After tmTNF-α or NTF was verified to express at transcriptional level in stably transfected Jurkat cells, the specificity of C1 to bind NTF was further confirmed by immunoprecipitation as well as confocal microscopy. Immunoprecipitation using C1, infliximab, or IgG was conducted followed by western blotting analysis. Although infliximab bound only tmTNF-α, C1 could bind either tmTNF-α or NTF, indicating that C1 specifically recognizes the NTF domain (Figure 1A). This finding was further verified by confocal microscopy analysis. Although tmTNF-α could be detected on the cell surface by either C1 or infliximab, NTFs on the cell surface were only recognized by C1 but not infliximab (Figure 1B).
Preferential expression of tmTNF-α in leukemia cells. (A) Lysates of Jurkat cells transfected with control, N-terminal truncated fragment (NTF), or tmTNF-α plasmid were precipitated with C1 and then were analyzed by western blotting. Infliximab and IgG served as controls. The image of immunoprecipitation (IP)-western blot is representative of 3 independent experiments. Infliximab bound only tmTNF-α protein but not NTF (middle panel). C1 could bind both tmTNF-α and NTF (black arrow) protein (right panel). Mouse IgG1 could not bind tmTNF-α or NTF protein (left panel). (B) tmTNF-α-negative Jurkat cells were transfected with control, NTF, or tmTNF-α plasmid and stained with fluorescence in situ hybridization, fluorescein isothiocyanate–conjugated C1 and phycoerythrin-conjugated infliximab. The image of confocal microscopy (magnification, ×1000) is representative of 3 independent experiments. (C) A scatter plot shows the percentages of tmTNF-α+ leukemia cells vs levels of serum sTNF-α in 18 patients with de novo AL. tmTNF-α expression was analyzed by flow cytometry using C1 in BM mononuclear cells from 69 AML, 30 ALL, and 30 nonmalignant donors (D), in CD34+CD38– or CD34+CD7–/CD19– fractions of AML, ALL, or nonmalignant cells (E), in CD34+CD7– (for T-ALL, n = 16) or CD34+CD19– (for B-ALL, n = 14) fractions of ALL (F), and in CD34+CD38– fractions among various French-American-British classification systems (FAB) AML subtypes (G). Data represent means ± SEM. ***P < .001 vs NBM (D) or HSC (E).
Preferential expression of tmTNF-α in leukemia cells. (A) Lysates of Jurkat cells transfected with control, N-terminal truncated fragment (NTF), or tmTNF-α plasmid were precipitated with C1 and then were analyzed by western blotting. Infliximab and IgG served as controls. The image of immunoprecipitation (IP)-western blot is representative of 3 independent experiments. Infliximab bound only tmTNF-α protein but not NTF (middle panel). C1 could bind both tmTNF-α and NTF (black arrow) protein (right panel). Mouse IgG1 could not bind tmTNF-α or NTF protein (left panel). (B) tmTNF-α-negative Jurkat cells were transfected with control, NTF, or tmTNF-α plasmid and stained with fluorescence in situ hybridization, fluorescein isothiocyanate–conjugated C1 and phycoerythrin-conjugated infliximab. The image of confocal microscopy (magnification, ×1000) is representative of 3 independent experiments. (C) A scatter plot shows the percentages of tmTNF-α+ leukemia cells vs levels of serum sTNF-α in 18 patients with de novo AL. tmTNF-α expression was analyzed by flow cytometry using C1 in BM mononuclear cells from 69 AML, 30 ALL, and 30 nonmalignant donors (D), in CD34+CD38– or CD34+CD7–/CD19– fractions of AML, ALL, or nonmalignant cells (E), in CD34+CD7– (for T-ALL, n = 16) or CD34+CD19– (for B-ALL, n = 14) fractions of ALL (F), and in CD34+CD38– fractions among various French-American-British classification systems (FAB) AML subtypes (G). Data represent means ± SEM. ***P < .001 vs NBM (D) or HSC (E).
Notably, levels of tmTNF-α did not significantly correlate with levels of serum sTNF-α in AL (Figure 1C), indicating that sTNF-α should not be considered to be a surrogate marker of tmTNF-α. Expression levels of tmTNF-α were significantly elevated in leukemia cells compared with nonmalignant mononuclear cells (P < .001; Figure 1D). We defined the hematopoietic stem cell (HSC)–enriched fractions as CD34+CD38– for both AML and nonmalignant donors. For B-ALL or T-ALL, the phenotypes of CD34+CD19– and CD34+CD7– were used, respectively, in the present study, although the existence, relevance, and exact phenotype of ALL leukemia stem cells (LSCs) are less clear.20 We found that the expression levels of tmTNF-α were much higher in HSC-enriched leukemia fractions than in nonmalignant cells (P < .001; Figure 1E). However, no significant differences in the levels of tmTNF-α expression could be observed among various subtypes of AL (Figure 1F-G). It is notable that within cells from a given leukemia patient, the expression of tmTNF-α in leukemia blasts correlated well with its expression in the HSC-enriched fraction (supplemental Figure 1G-H).
Levels of tmTNF-α expression were compared between cases of AML and ALL according to various clinical parameters. The expression levels of tmTNF-α appeared to correlate with the adverse clinical features of AL. In AML, high levels of tmTNF-α expression were significantly correlated with a higher percentage of CD34+ cells, extramedullary infiltration, and an adverse risk group (Table 1). In ALL, high tmTNF-α expression levels were significantly correlated with a higher white blood cell count, extramedullary infiltration, and a poor risk group (Table 2).
Expression of tmTNF-α facilitates the survival of AL cells by modulating multiple signaling pathways
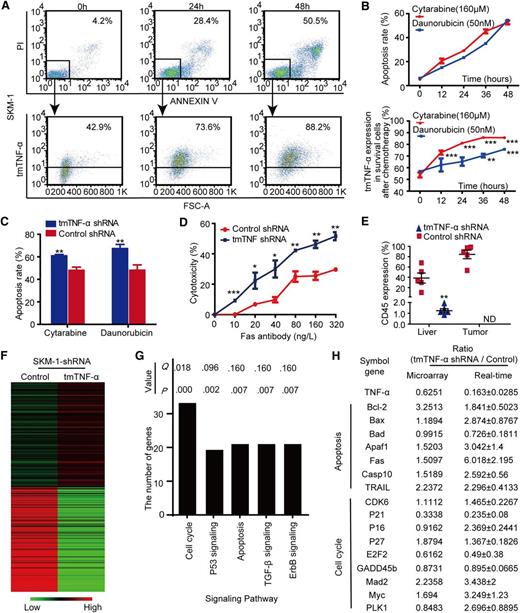
SKM-1 and HL-60 cells were exposed to a series of concentrations of chemotherapeutic agents for 48 hours to determine dosages that induced ∼50% apoptosis (in red), and these empirically determined doses were then selected to treat cells for different amounts of time (supplemental Figure 2A-B). Surviving leukemia cells were gated and assessed for the expression of tmTNF-α (Figure 2A and supplemental Figure 2C). We found that tmTNF-α+ cells were significantly enriched in surviving leukemia cells in a time-dependent fashion after exposure to cytarabine or daunorubicin and gave rise to a sharp increase in tmTNF-α expression in residual leukemia cells (Figure 2B and supplemental Figure 2D). These findings suggested that knockdown of tmTNF-α expression may render leukemia cells more sensitive to chemotherapy.
Expression of tmTNF-α facilitates the survival of AML cells by modulating multiple signaling pathways. (A-B) SKM-1 cells were treated with 160 μM cytarabine and 50 nM daunorubicin for indicated time points. The apoptosis was detected by staining with propidium iodide (PI) and Annexin-V (B, upper panel), and the double negative cells were gated as surviving leukemia cells after chemotherapy (A, upper panel). tmTNF-α expression was determined in these cells by flow cytometry (A and B, lower panel). Each flow cytogram is representative of 3 independent experiments. The quantitative data represent the mean ± SEM of at least 3 independent experiments. **P < .01; ***P < .001 vs the 0 hours group. (C) Each bar represents the mean ± SEM of the apoptosis rate in control or tmTNF-α shRNA-transfected SKM-1 cells treated by either cytarabine or daunorubicin for 48 hours; **P < .01 vs control shRNA. (D) Each curve represents the mean ± SEM of the cytotoxicity rate of control or tmTNF-α shRNA-transfected SKM-1 cells after treatment with agonistic anti-Fas antibody for 24 hours; *P < .05; ** P < .01; ***P < .001 vs control shRNA-transfection group. (E) Control or tmTNF-α shRNA-transfected SKM-1 cells (1 × 106) were transplanted into recipient NOD-SCID mice to initiate leukemia (n = 5). Recipient mice were euthanized after 6 weeks. At the experimental end point, the percentages of CD45+ cells in the murine liver and in palpable tumors were determined by flow cytometry. Data are presented as scatter dot plots with means (horizontal lines); **P < .01 vs the control shRNA-transfected group; ND, FCM assessment was not performed because no mice in the tmTNF-α shRNA group developed any palpable tumors. (F) A heatmap shows gene expression profiling associated with control- or tmTNF-α shRNA-transfected SKM-1 leukemia cells. (G) The most significantly enriched pathways showing differential expression between the 2 groups were identified by DAVID pathway analysis. Each column represents the number of differentially expressed genes between the 2 groups. For differentially expressed genes, the call value of expression had to be P < .05, with a ratio ≥1.5 or ≤0.6667. Both Q and P values are shown. (H) Selected genes that were differentially expressed between the 2 groups were confirmed by qPCR analysis. Values indicate the mean expression ratio comparing the tmTNF-α-knockdown group with the control group.
Expression of tmTNF-α facilitates the survival of AML cells by modulating multiple signaling pathways. (A-B) SKM-1 cells were treated with 160 μM cytarabine and 50 nM daunorubicin for indicated time points. The apoptosis was detected by staining with propidium iodide (PI) and Annexin-V (B, upper panel), and the double negative cells were gated as surviving leukemia cells after chemotherapy (A, upper panel). tmTNF-α expression was determined in these cells by flow cytometry (A and B, lower panel). Each flow cytogram is representative of 3 independent experiments. The quantitative data represent the mean ± SEM of at least 3 independent experiments. **P < .01; ***P < .001 vs the 0 hours group. (C) Each bar represents the mean ± SEM of the apoptosis rate in control or tmTNF-α shRNA-transfected SKM-1 cells treated by either cytarabine or daunorubicin for 48 hours; **P < .01 vs control shRNA. (D) Each curve represents the mean ± SEM of the cytotoxicity rate of control or tmTNF-α shRNA-transfected SKM-1 cells after treatment with agonistic anti-Fas antibody for 24 hours; *P < .05; ** P < .01; ***P < .001 vs control shRNA-transfection group. (E) Control or tmTNF-α shRNA-transfected SKM-1 cells (1 × 106) were transplanted into recipient NOD-SCID mice to initiate leukemia (n = 5). Recipient mice were euthanized after 6 weeks. At the experimental end point, the percentages of CD45+ cells in the murine liver and in palpable tumors were determined by flow cytometry. Data are presented as scatter dot plots with means (horizontal lines); **P < .01 vs the control shRNA-transfected group; ND, FCM assessment was not performed because no mice in the tmTNF-α shRNA group developed any palpable tumors. (F) A heatmap shows gene expression profiling associated with control- or tmTNF-α shRNA-transfected SKM-1 leukemia cells. (G) The most significantly enriched pathways showing differential expression between the 2 groups were identified by DAVID pathway analysis. Each column represents the number of differentially expressed genes between the 2 groups. For differentially expressed genes, the call value of expression had to be P < .05, with a ratio ≥1.5 or ≤0.6667. Both Q and P values are shown. (H) Selected genes that were differentially expressed between the 2 groups were confirmed by qPCR analysis. Values indicate the mean expression ratio comparing the tmTNF-α-knockdown group with the control group.
To further assess the impact of tmTNF-α expression on leukemia cells, SKM-1 cells were stably transfected with tmTNF-α shRNA, and knockdown of the messenger RNA was confirmed (supplemental Figure 2E). When tmTNF-α shRNA-transfected SKM-1 cells were exposed to cytarabine or daunorubicin for 48 hours, knockdown of tmTNF-α led to significantly increased apoptosis or cytotoxicity induced either by chemotherapy (Figure 2C) or by Fas agonistic antibody21 (Figure 2D), indicating that knockdown of the tmTNF-α expression renders leukemia cells more sensitive to proapoptotic stimuli. Furthermore, silence of tmTNF-α expression resulted in markedly fewer colonies compared with control cells (P = .0058), showing a decreased clonogenic potential in the leukemia cells (supplemental Figure 2F). In vivo, all mice transplanted with control shRNA-transfected SKM-1 cells developed signs of AML, including decreased activity, AML cell infiltration in the liver, and the formation of palpable tumors, although leukemia did not develop in the peripheral blood (PB), BM, or spleen. Surprisingly, knockdown of tmTNF-α in SKM-1 cells failed to regenerate leukemia in recipient mice at the end of the experiment, indicating that tmTNF-α expression is essential for the repopulation of the AML cells in NOD-SCID mice (Figure 2E).
To understand the potential mechanisms underlying the effects caused by tmTNF-α knockdown, microarray analysis was carried out (Figure 2F). More than 3000 differentially expressed genes were identified in SKM-1 cells that were stably transfected with tmTNF-α shRNA compared with control cells. DAVID pathway analysis revealed that the knockdown of tmTNF-α gave rise to a substantial change in multiple signaling pathways, and the pathways with the most differentially expressed genes included the cell cycle and apoptosis pathways (Figure 2G). Expression patterns of selected genes from the microarray data set were further validated by qPCR. For the apoptosis pathway, tmTNF-α knockdown yielded a total of 19 differentially expressed genes, including 12 proapoptotic genes (Figure 2H). For the cell cycle pathway, there were a total of 28 differentially expressed genes, including 16 genes that inhibited cell cycle progression (Figure 2H). We further examined the protein expression patterns of selected genes based on microarray data (supplemental Figure 2G). Notably, shRNA-mediated tmTNF-α knockdown significantly increased the protein expression of the critical proapoptotic regulators Apaf1, Bax, Fas, and TRAIL in SKM-1 cells. Additionally, protein expression levels of the critical negative cell cycle regulators P16 and P27 were enhanced in SKM-1 cells that were stably transfected with tmTNF-α shRNA. These data indicated that tmTNF-α knockdown favors both the induction of apoptosis and the inhibition of cell cycle progression. To further explore the clinical relevance of gene expression profile data derived from SKM-1 cells, a primary AML sample in which tmTNF-α expression was 14.6% (supplemental Figure 2H) was sorted into tmTNF-α+ and tmTNF-α– fractions, which were then subjected to microarray analysis. Analysis of microarray data showed that the global gene expression patterns in tmTNF-α+ leukemia cells were substantially different from those in tmTNF-α– leukemia cells (supplemental Figure 2I). More than 4000 differentially expressed genes were identified between the different cell fractions. DAVID pathway analysis revealed that the pathways with the most differentially expressed genes included the cell cycle, apoptosis, and P53 signaling pathways (supplemental Figure 2J). For the apoptosis pathway, tmTNF-α− leukemia cells induced a total of 29 differentially expressed genes, including 15 proapoptotic genes. For the cell cycle pathway, there were a total of 54 differentially expressed genes, including 29 genes associated with the inhibition of cell cycle progression. In accord with our findings in SKM-1 cells, primary tmTNF-α− leukemia cells showed a gene expression profile that favored the induction of apoptosis and inhibition of cell cycle progression (supplemental Table 4).
Expression profiles of tmTNF-α in normal hematopoietic cells and nonhematopoietic tissues
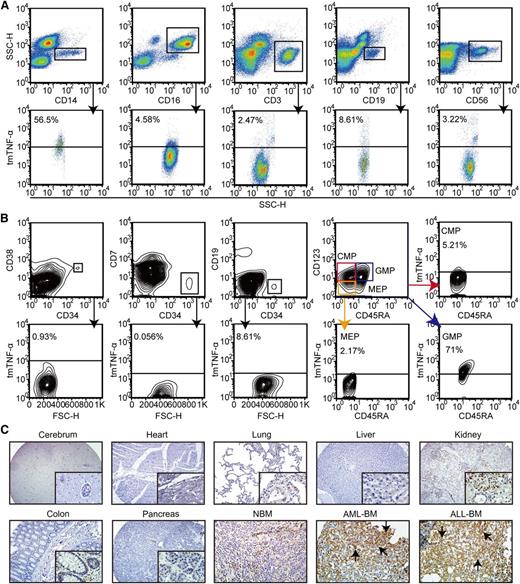
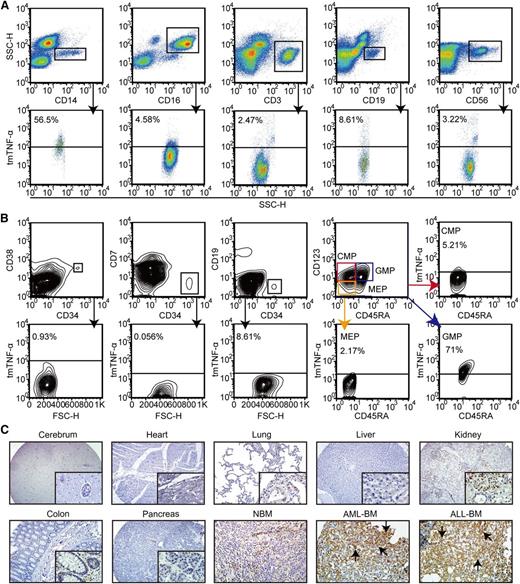
Within PB leukocytes, C1 mAb specifically recognized 56.5% of CD14+ monocytes and 71% of granulocyte/monocyte progenitors.22 A trace amount of tmTNF-α expression could be observed in CD16+ granulocytes, CD3+ T cells, CD19+ B cells, and CD56+ NK cells, as well as NBM and normal hematopoietic stem/progenitor cells (Figure 3A-B and supplemental Figure 3A).22-24 Strongly positive tmTNF-α staining was detected in both AML and ALL specimens but was weakly positive or negative in normal human tissues and NBM biopsy tissues (Figure 3C and supplemental Figure 3B).
Analysis of tmTNF-α expression in normal human blood cells and tissues. (A) Expression of tmTNF-α by various types of white blood cells in PB from healthy donors (n = 4) was analyzed by FCM. The types of white blood cells were gated using the indicated antibodies. (B) Various lineages of hematopoietic stem/progenitor cells in healthy BM (n = 5) were gated using the indicated antibodies and assessed for tmTNF-α expression. (C) Expression of tmTNF-α was examined by immunohistochemical staining of histology sections from various normal (magnification, ×100 and ×400) and malignant human tissues (magnification, ×400). Immunoreactive tmTNF-α (black arrows) was readily detectable in BM sections with de novo AML or ALL, but was absent or only weakly detected in normal human tissues (2 sections per tissue for each tissue microarray).
Analysis of tmTNF-α expression in normal human blood cells and tissues. (A) Expression of tmTNF-α by various types of white blood cells in PB from healthy donors (n = 4) was analyzed by FCM. The types of white blood cells were gated using the indicated antibodies. (B) Various lineages of hematopoietic stem/progenitor cells in healthy BM (n = 5) were gated using the indicated antibodies and assessed for tmTNF-α expression. (C) Expression of tmTNF-α was examined by immunohistochemical staining of histology sections from various normal (magnification, ×100 and ×400) and malignant human tissues (magnification, ×400). Immunoreactive tmTNF-α (black arrows) was readily detectable in BM sections with de novo AML or ALL, but was absent or only weakly detected in normal human tissues (2 sections per tissue for each tissue microarray).
Administration of tmTNF-α antibody effectively kills leukemia cells through CDC and ADCC in vitro
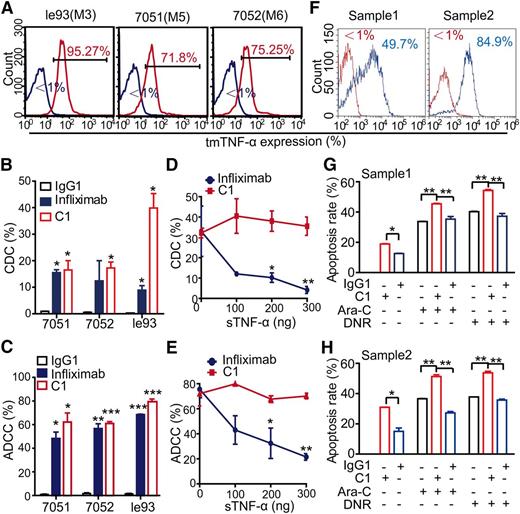
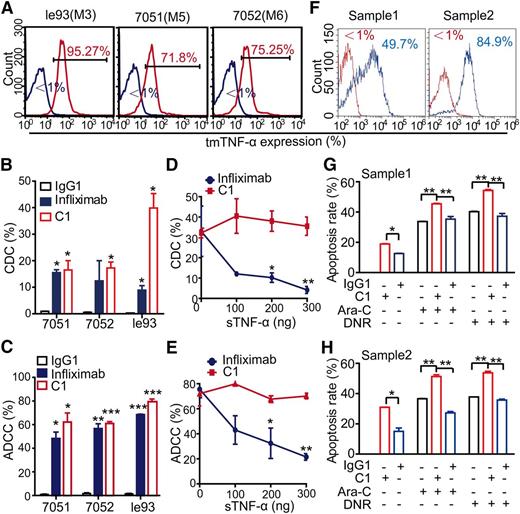
The le93, 7051, and 7052 cell lines were representative of AML-M3, -M5, and -M6, respectively, and all of these cell lines expressed high levels of tmTNF-α (Figure 4A). In the presence of complement or immune effector cells, either infliximab or C1 antibodies showed complement dependent cytotoxicity (CDC) and antibody-dependent cell-mediated cytotoxicity (ADCC) against all 3 types of leukemia cells, but C1 antibody displayed similar or more potent activities than infliximab (Figure 4B-C). The isotype control antibody showed no killing effects under similar conditions. Moreover, infliximab-mediated CDC and ADCC were mostly abolished by the addition of sTNF-α in a dose-dependent manner; however, the 2 cytotoxic activities of C1 remained unaffected in the presence of sTNF-α (Figure 4D-E). To address whether treatment with C1 sensitized leukemia cells to chemotherapy, 2 primary tmTNF-α+ AML samples were selected (Figure 4F). CDC assays was initially performed on primary AML cells, followed by treatment with cytarabine or daunorubicin. As expected, treatment with C1 could significantly sensitize leukemia cells to chemotherapy (Figure 4G-H).
Anti-tmTNF-α antibody effectively kills leukemia cells via CDC and ADCC. (A) Expression of tmTNF-α by 3 leukemia cells lines was determined by FCM. The FCM plots are representative of 3 independent experiments. Mouse IgG1 was used as an isotype control. (B-C) The CDC and ADCC activity of C1 and infliximab. For the CDC assay (B), leukemia cells were incubated with 2 μg/mL C1, infliximab, or isotype control IgG1 in the presence of either guinea pig complement (for C1 or IgG1) or human complement (for infliximab) at 37°C for 2 hours. For the ADCC assay (C), leukemia cells were incubated with 2 μg/mL of the indicated antibodies in the presence of either murine macrophages (for C1 or IgG1) or human PBMCs (for infliximab) at 37°C for 4 hours. Data are expressed as means ± SEM (n = 3); *P < .05; **P < .01; ***P < .001 vs the isotype control group. (D-E) The CDC and ADCC activities of C1 and infliximab were determined in the presence of increasing concentrations of sTNF-α. The CDC (D) and ADCC (E) activity of infliximab, but not C1, could be significantly neutralized by the addition of sTNF-α. (F) Expression of tmTNF-α on 2 primary AML cells was determined by FCM. As an isotype control, mouse IgG1 was used. (G-H) Leukemia cells were incubated with 2 μg/mL C1 or isotype control IgG1 in the presence of guinea pig complement for 2 hours, followed by treatment with either cytarabine (160 μmol/L) or daunorubicin (50 nmol/L) at 37°C for an additional 24 hours. The apoptosis was determined in triplicate by staining with Annexin V and PI. Data are expressed as means ± SEM; *P < .05; **P < .01 vs the isotype control group.
Anti-tmTNF-α antibody effectively kills leukemia cells via CDC and ADCC. (A) Expression of tmTNF-α by 3 leukemia cells lines was determined by FCM. The FCM plots are representative of 3 independent experiments. Mouse IgG1 was used as an isotype control. (B-C) The CDC and ADCC activity of C1 and infliximab. For the CDC assay (B), leukemia cells were incubated with 2 μg/mL C1, infliximab, or isotype control IgG1 in the presence of either guinea pig complement (for C1 or IgG1) or human complement (for infliximab) at 37°C for 2 hours. For the ADCC assay (C), leukemia cells were incubated with 2 μg/mL of the indicated antibodies in the presence of either murine macrophages (for C1 or IgG1) or human PBMCs (for infliximab) at 37°C for 4 hours. Data are expressed as means ± SEM (n = 3); *P < .05; **P < .01; ***P < .001 vs the isotype control group. (D-E) The CDC and ADCC activities of C1 and infliximab were determined in the presence of increasing concentrations of sTNF-α. The CDC (D) and ADCC (E) activity of infliximab, but not C1, could be significantly neutralized by the addition of sTNF-α. (F) Expression of tmTNF-α on 2 primary AML cells was determined by FCM. As an isotype control, mouse IgG1 was used. (G-H) Leukemia cells were incubated with 2 μg/mL C1 or isotype control IgG1 in the presence of guinea pig complement for 2 hours, followed by treatment with either cytarabine (160 μmol/L) or daunorubicin (50 nmol/L) at 37°C for an additional 24 hours. The apoptosis was determined in triplicate by staining with Annexin V and PI. Data are expressed as means ± SEM; *P < .05; **P < .01 vs the isotype control group.
Direct in vivo administration of tmTNF-α antibody selectively inhibited AML cell growth in NOD-SCID mice
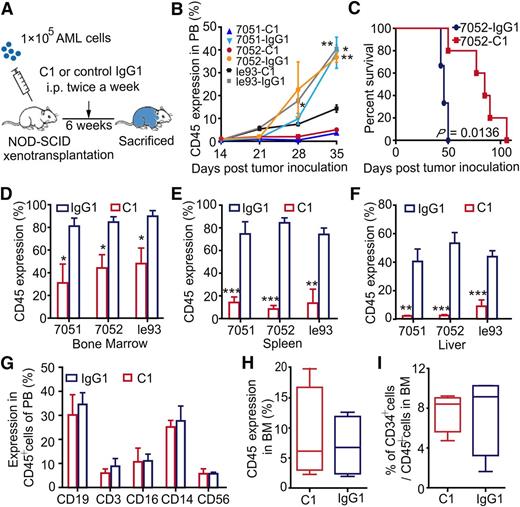
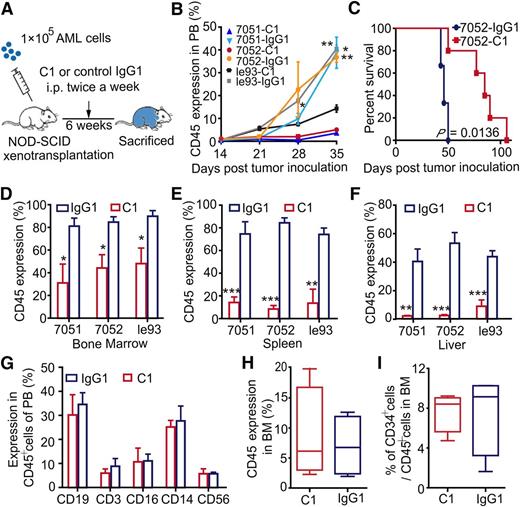
Leukemia cells were transplanted into NOD-SCID mice to regenerate AML hematopoiesis. C1 treatment was initiated 24 hours posttransplantation and administered twice per week for a total of 6 weeks (Figure 5A). After administration of C1, concentrations of C1 rapidly reached a steady serum concentration (supplemental Figure 5A). C1 treatment not only significantly delayed the appearance of AML cells in PB compared with IgG1 control antibody (Figure 5B), but also resulted in markedly improved survival (P = .0136; Figure 5C). AML engraftment in the BM (Figure 5D), spleen (Figure 5E), and liver (Figure 5F) was consistently impaired in the C1 group. Meanwhile, spleen weights were lower in the C1 group than in the IgG1 control group (supplemental Figure 5B), indicating that C1 administration efficiently reduced the AML burden in NOD-SCID mice.
In vivo administration of tmTNF-α antibody inhibits AML cell growth in NOD-SCID mice but does not impair the repopulation capacity of normal hematopoietic cells. (A) An overview of the experimental design: each NOD-SCID recipient mouse was transplanted with 1 × 105 leukemia cells. Treatment with C1 or IgG1 control antibody (n = 5) was initiated at day 1 posttransplantation. Antibodies were administered intraperitoneally at 10 μg per 1 g body weight twice per week for 6 weeks; mice were euthanized at the end of the 6-week period. (B) The percentage of human AML cells in PB of mice. Values represent means ± SEM; *P < .05; **P < .01 between IgG1 control and C1-treated groups. (C) Kaplan-Meier survival curves of NOD-SCID mice treated with C1 or IgG1 control antibody. For the survival curve analysis, twice weekly antibody treatment was continued until a mouse died. Survival curves were compared using the log-rank test. Error bars represent means ± SEM; *P < .05 between control and C1-treated groups. (D-F) Percentage of human AML cells in murine BM (D), spleen (E), or liver (F) at the end of the experiment, as assessed by FCM. Error bars represent means ± SEM for the percentage of leukemia cells in each group. (G-I) Each NOD-SCID recipient mouse was transplanted with 1 × 105 CD34+ cells harvested from the BM of healthy human donors. Treatment with C1 or IgG1 control antibody (n = 4 each group) was administered following the same protocol described previously. The effects of C1 on normal hematopoietic cells were assessed at the experimental end point. (G) The proportion of various types of human blood cells in PB of mice from the C1 or IgG1 control group. Human engraftment in mouse BM (H) and the percentage of CD34+ cells in human grafts within mouse BM (I) are shown. Error bars represent means ± SEM in the C1 vs IgG1 groups; no significant difference was detected between the groups.
In vivo administration of tmTNF-α antibody inhibits AML cell growth in NOD-SCID mice but does not impair the repopulation capacity of normal hematopoietic cells. (A) An overview of the experimental design: each NOD-SCID recipient mouse was transplanted with 1 × 105 leukemia cells. Treatment with C1 or IgG1 control antibody (n = 5) was initiated at day 1 posttransplantation. Antibodies were administered intraperitoneally at 10 μg per 1 g body weight twice per week for 6 weeks; mice were euthanized at the end of the 6-week period. (B) The percentage of human AML cells in PB of mice. Values represent means ± SEM; *P < .05; **P < .01 between IgG1 control and C1-treated groups. (C) Kaplan-Meier survival curves of NOD-SCID mice treated with C1 or IgG1 control antibody. For the survival curve analysis, twice weekly antibody treatment was continued until a mouse died. Survival curves were compared using the log-rank test. Error bars represent means ± SEM; *P < .05 between control and C1-treated groups. (D-F) Percentage of human AML cells in murine BM (D), spleen (E), or liver (F) at the end of the experiment, as assessed by FCM. Error bars represent means ± SEM for the percentage of leukemia cells in each group. (G-I) Each NOD-SCID recipient mouse was transplanted with 1 × 105 CD34+ cells harvested from the BM of healthy human donors. Treatment with C1 or IgG1 control antibody (n = 4 each group) was administered following the same protocol described previously. The effects of C1 on normal hematopoietic cells were assessed at the experimental end point. (G) The proportion of various types of human blood cells in PB of mice from the C1 or IgG1 control group. Human engraftment in mouse BM (H) and the percentage of CD34+ cells in human grafts within mouse BM (I) are shown. Error bars represent means ± SEM in the C1 vs IgG1 groups; no significant difference was detected between the groups.
Next, we investigated the sensitivity of NBM to C1 treatment using the same strategy as for AML cells. In PB, human CD14+ monocyte/macrophage cell counts were slightly, but not significantly reduced in the C1 group. Counts of human CD16+ granulocytes, CD19+ B cells, CD3+ T cells, and CD56+ NK cells were similar between the 2 groups (Figure 5G). Moreover, no inhibitory effect of C1 on the engraftment of normal human CD45+ cells in NOD-SCID mouse BM (Figure 5H) and spleen (supplemental Figure 5C) was observed. The frequencies of CD34+ cells within the human engrafted cells were not reduced by C1 treatment (Figure 5I). The histologic morphology of liver, kidney, heart, and lung tissues were analyzed by hematoxylin and eosin staining and did not reveal differences between the C1- and IgG1- treated groups (supplemental Figure 5D).
Treatment with tmTNF-α antibody targets LSCs in vivo
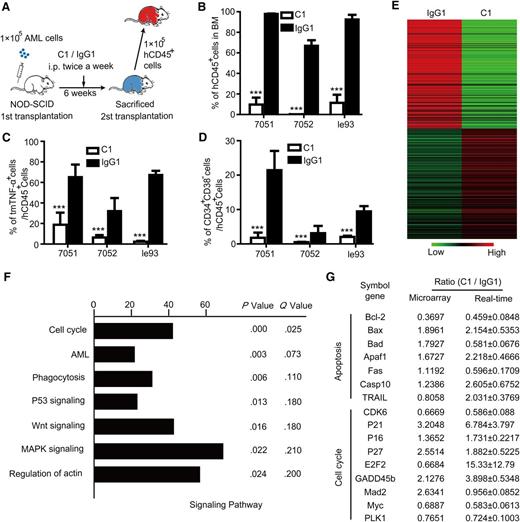
To determine whether C1 treatment affects LSCs, a second serial transplantation was performed using CD45+ leukemia cells derived from C1- or IgG1- treated NOD-SCID mice after the first transplantation.22 Each NOD-SCID recipient mouse was transplanted with 1 × 105 CD45+ cells, and leukemia was allowed to regenerate for 6 weeks without any further treatment (Figure 6A). Surprisingly, the appearance of hCD45+ cells was significantly delayed in the PB of the C1 group as compared with that of the IgG1 control group (supplemental Figure 6A). Furthermore, human AML engraftment in the BM and spleen was significantly attenuated in the C1 group as compared with that of the IgG1 control group (Figure 6B and supplemental Figure 6B). Notably, C1 treatment during the first transplantation was sufficient to result in the dramatic depletion of both tmTNF-α+ and CD34+ AML cells among total engrafted leukemia cells in the secondary transplant recipient mice (Figure 6C-D), indicating that LSCs were impaired by C1 treatment.
Treatment with tmTNF-α antibody targets LSCs in vivo. (A) An overview of the experimental design. At the end of the first transplantation, leukemia cells were harvested. For the second transplantation, each NOD-SCID recipient mouse was transplanted with 1 × 105 CD45+ cells from C1- or IgG1- treated mice (n = 5 each group). Engraftment of AML cells in NOD-SCID mice was determined 6 weeks posttransplantation. (B) Levels of AML cell engraftment into the BM of secondary recipient mice were determined by FCM. Vertical bars represent means ± SEM; ***P < .001 between control and C1- treated groups. (C) The percentage of tmTNF-α+ cells within the total engrafted AML cells of secondary recipient mice was determined by FCM; ***P < .001 between control and tmTNF-α-treated groups. (D) The percentage of CD34+ AML cells among total engrafted AML cells in the BM of secondary recipient mice was determined by FCM. Values represent means ± SEM; ***P < .001 between the control and C1-treated groups. (E) Heatmap of gene expression profiles associated with leukemia cells harvested from control or C1- treated groups at the end of the first transplantation. (F) The most highly enriched pathways exhibiting differential expression between the control- and C1- treated groups were identified using DAVID pathway analysis. Each column indicates the number of differentially expressed genes between the 2 groups. For the differentially expressed genes, the call value of expression had to be P < .05, with an expression ratio ≥1.3 or ≤0.7692. Both Q and P values are shown. (G) Typical genes that are differentially expressed between control- and C1- treated groups that were confirmed by qPCR. Values represent mean expression ratios comparing the C1- treated and control groups.
Treatment with tmTNF-α antibody targets LSCs in vivo. (A) An overview of the experimental design. At the end of the first transplantation, leukemia cells were harvested. For the second transplantation, each NOD-SCID recipient mouse was transplanted with 1 × 105 CD45+ cells from C1- or IgG1- treated mice (n = 5 each group). Engraftment of AML cells in NOD-SCID mice was determined 6 weeks posttransplantation. (B) Levels of AML cell engraftment into the BM of secondary recipient mice were determined by FCM. Vertical bars represent means ± SEM; ***P < .001 between control and C1- treated groups. (C) The percentage of tmTNF-α+ cells within the total engrafted AML cells of secondary recipient mice was determined by FCM; ***P < .001 between control and tmTNF-α-treated groups. (D) The percentage of CD34+ AML cells among total engrafted AML cells in the BM of secondary recipient mice was determined by FCM. Values represent means ± SEM; ***P < .001 between the control and C1-treated groups. (E) Heatmap of gene expression profiles associated with leukemia cells harvested from control or C1- treated groups at the end of the first transplantation. (F) The most highly enriched pathways exhibiting differential expression between the control- and C1- treated groups were identified using DAVID pathway analysis. Each column indicates the number of differentially expressed genes between the 2 groups. For the differentially expressed genes, the call value of expression had to be P < .05, with an expression ratio ≥1.3 or ≤0.7692. Both Q and P values are shown. (G) Typical genes that are differentially expressed between control- and C1- treated groups that were confirmed by qPCR. Values represent mean expression ratios comparing the C1- treated and control groups.
To explore the potential mechanisms underlying the therapeutic actions of C1, we assessed cellular homing efficiency and found that there was not significantly different between the 2 groups (supplemental Figure 6C). Next, CD45+ leukemia cells were collected from the first recipient mice and compared for the global gene expression profiling experiment. Microarray data revealed that the overall gene expression patterns in the C1 and IgG1 control groups were markedly different (Figure 6E), and we identified >5000 differentially expressed genes between the 2 groups. DAVID pathway analysis showed that the most differentially affected pathways included the cell cycle, P53, and WNT signaling pathways (Figure 6F). Selected genes identified by microarray analysis were further validated by qPCR (Figure 6G). In accord with our previous report, actin cytoskeleton association with the tmTNF-α pathway had been identified as one of the most differentially expressed pathways.25 Although our DAVID pathway analysis had failed to enrich apoptosis as the most differential pathways, we still examined the apoptosis pathway based on our findings in vitro experiments (Figure 2H). Treatment with C1 resulted in a total of 22 differentially expressed genes, including 13 genes that favored the induction of apoptosis. In line with our findings in vitro, 13 critical proapoptotic genes, including Apaf1, Bad, Bax, Bcl-2, Fas and TRAIL are substantially affected at the transcriptional level. On the other hand, C1 treatment also led to 9 differentially expressed genes that exerted antiapoptotic activities, which were predicted to yield protective responses in the surviving leukemia cells. For the cell cycle pathway, C1 treatment resulted in 31 differentially expressed genes, including 21 genes that favored the suppression of cell cycle progression. Strikingly, the expression of many key cell cycle regulators, such as P16, P21, P27, GADD45b, E2F2, PLK1, and Myc, were substantially affected by C1 treatment (Figure 6G).
Discussion
The identification of novel cell surface antigens is essential for the design of effective antibody therapies to eliminate LSCs. Targeting cell surface antigens that exhibit druggable features and also play critical roles in leukemia maintenance represents an attractive strategy. Herein, using the C1 that specifically recognizes the tmTNF-α domain without cross-reacting with sTNF-α, we can show a favorable AL-associated expression profile of tmTNF-α that is important for AL cell survival and proliferation. Notably, the expression levels of tmTNF-α did not correlate with serum sTNF-α concentrations in AL, indicating that serum sTNF-α does not represent a surrogate marker for tmTNF-α, as we and others expected. Notably, tmTNF-α was preferentially expressed by AL blasts and within an LSC-enriched subpopulation. Clinically, although we did not have enough cases of patient cohort at this moment to conclude whether tmTNF-α is an independent prognostic indicator in a stepwise regression multivariate analysis, high tmTNF-α expression was associated with both poor risk stratification and adverse clinical parameters, such as the extramedullary infiltration of leukemia and high white blood counts. Apparently, whether tmTNF-α is an independent prognostic indicator will be a critical and interesting issue to be addressed in future study. In vitro, knockdown of tmTNF-α expression rendered leukemia cells more sensitive to chemotherapy and Fas-induce cytotoxicity and decreased clonogenic potential. In vivo, silence of tmTNF-α expression in SKM-1 cells failed to regenerate leukemia in recipient mice, indicating that tmTNF-α expression is essential for the repopulation of the AML cells in NOD-SCID mice. These findings highlight the great potential for exploiting tmTNF-α as a therapeutic target to eradicate LSCs and leukemia blasts in future AL therapies.
Targeting tmTNF-α by C1 antibody in leukemia represents an attractive approach for the following reasons.26 First, the AL-associated expression pattern of tmTNF-α should allow for maximal antileukemia activity and minimal off-target toxicity to normal tissues. Second, a markedly higher expression level of tmTNF-α was consistently detected in LSCs, but not in normal HSCs, which presents an opportunity for the selective elimination of leukemia stem/progenitor cells that would be otherwise resistant to conventional chemotherapy. Third, the expression of tmTNF-α itself was critical for the repopulation and growth of AML cells in vivo because it affected multiple signaling pathways. These results are in accord with previous publications and underscore the importance of TNF-α in maintaining leukemia hematopoiesis.27,28 Finally, the C1 was designed to recognize the NTF that can remain on the cell surface after tmTNF-α cleavage by TNF-α-converting enzyme.14 Therefore, this antibody can recognize both tmTNF-α (full-length)-expressing and sTNF-α-secreting cells.
Our findings demonstrated that C1 could kill tmTNF-α+ leukemia cells in vitro via ADCC and CDC, which is consistent with our previous report showing these activities of C1 in a tmTNF-α+ breast cancer cell line.14 Meanwhile, the effects of a combination of chemotherapeutics with C1 indicated that treatment with C1 could sensitize leukemia cells to chemotherapy. Additionally, C1 not only selectively inhibited tmTNF-α+ AML cell growth in NOD-SCID mice without affecting NBM transplantation, but also depleted tmTNF-α+ AML and CD34+ cells in secondary recipient mice, suggesting that C1 antibody markedly reduced both the leukemia burden and capacity to regenerate AML, indicating the effective targeting of LSCs by this antibody. As tmTNF-α+ leukemia cells appeared to be resistant subclones that could be highly enriched by chemotherapy, the selective eradication of tmTNF-α+ leukemia subclones by C1 resulted in a less aggressive residual leukemia population with a signature of indolent cell cycle progression and more susceptibility to apoptosis, as revealed by microarray analysis. These findings yield insights into the mechanism of action for C1 and further underscore the clinical potential and relevance of tmTNF-α in AL in avoiding the enrichment of refractory leukemia subclones by chemotherapy.
Together, our findings indicate that tmTNF-α is a key protumor molecule for AL and a potent novel target for both AL and LSCs. Targeting tmTNF-α by C1 antibody can effectively eradicate AL cells and LSCs, thereby preventing disease relapse. Moreover, considering that proliferation, survival, and other malignant features of tumor cells can be initiated and maintained by sTNF-α-related inflammation,29-32 the ability of C1 to target TNF-α-producing sources is expected to curtail and possibly even reverse sTNF-α-related supportive microenvironments in a way that extends beyond the killing of AL blasts and LSCs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Crown BioScience Incorporated Company (Beijing) for technique assistance in the NOD-SCID mice study.
This work was supported by grants from the National Science Foundation for Young Scholars of China (81300415, X.Z.), the National Natural Science Funds for Distinguished Young Scholar (grant 81025011, J.Z.), and the Key Program of National Natural Science Foundation of China (grants 81230052 and 81090414, J.Z.).
Authorship
Contribution: J.Z. and Z.L. designed and coordinated the study; X.Z. and S.Z. analyzed the clinical samples, completed the animal experiments and partial experiments in vitro, performed the statistical analysis, and wrote the manuscript; B.L. generated the mAb C1 used in this study; and all coauthors contributed to the clinical sample collecting and the experiments in vitro, supported the study conduct, and contributed to the final report.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jianfeng Zhou, Tongji Hospital, 1095# Jie-Fang-Da-Dao, Wuhan 430030, China; e-mail: jfzhou@tjh.tjmu.edu.cn; Zhuoya Li, Tongji Medical College, Huazhong University of Science and Technology, 13 Hangkong Rd, Wuhan 430030, China; e-mail: zhuoyali@mails.tjmu.edu.cn.
References
Author notes
X.Z. and S.Z. contributed equally to this study.