To the editor:
Multiple myeloma is often treated with high-dose melphalan chemotherapy followed by maintenance therapy with immunomodulatory agents aimed to delay disease progression and prolong survival. Although it is routinely offered to many patients, randomized trials of maintenance therapy using thalidomide or lenalidomide only demonstrated improvement in progression-free survival (PFS), with inconsistent overall survival (OS) benefit.1-4 Genetic biomarkers predictive of the efficacy and toxicity of immunomodulatory drug maintenance therapy, as well as prognosis following front-line high-dose melphalan therapy, are of potential value in identifying patients best suited for maintenance therapy. Cibeira et al found that single-nucleotide polymorphisms (SNPs) in DNA repair genes were associated with thalidomide response and survival outcome.5 Johnson et al found several SNP associations with risk of venous thrombosis6 and thalidomide-related peripheral neuropathy,7 respectively. Dumontet et al found that SNPs in drug metabolism and DNA repair genes were associated with response to high-dose melphalan and survival.8 Fernández de Larrea et al found that SNPs in FAM179B and MIR196A were associated with survival after lenalidomide salvage therapy.9 Well-designed replication studies with independent patient cohorts are crucial in pharmacogenetics to facilitate clinical uptake of predictive biomarkers. In addition, randomized trial design is required for rigorous evaluation of predictive markers associated with benefit or harm from therapy, where the interaction between treatment assignment and outcome can be examined; otherwise, a prognostic marker can be mistakenly identified as predictive.10
In our study, 44 validation SNPs were selected from previous studies with significant associations with myeloma prognosis and/or thalidomide and melphalan treatment efficacy and toxicity5-9,11 for validation in the Canadian Cancer Trials Group (formerly named the National Cancer Institute of Canada Clinical Trials Group [NCIC CTG]) MY.10 randomized trial. Fourteen exploratory SNPs in genes shown to be involved in myeloma disease pathways as well as the CRBN-IKZF1/IKZF3-IRF4 pathway,12-19 which is now known to be central to the mechanism of action of immunomodulatory drugs, were also investigated (the full list of candidate SNPs can be found in supplemental Table 1, see supplemental Data available at the Blood Web site). Exploratory SNPs were chosen due to known functional significance or had clinically significant impact in other disease settings. The MY.10 randomized trial compared thalidomide-prednisone maintenance therapy with observation post high-dose melphalan chemotherapy and autologous stem cell transplant (ASCT).20 Bone marrow aspirates (n = 86 treatment arm; n = 101 observation arm) were obtained (for patient demographic details, see supplemental Table 2). Genomic DNA were extracted and SNP genotypes were assessed by allelic discrimination using TaqMan assays on the ViiA7 platform. To rule out potential discrepancies in SNP genotype between tumor and germ line DNA in our candidate SNPs, concordance assays between tumor and germ line DNA extracted from paired bone marrow and blood samples, respectively, from 9 patients in a separate cohort were conducted and found to be completely concordant for all 58 SNPs. Cox regression models were performed using SAS (version 9.3) for prognostic impact of a single SNP on PFS and OS, with and without adjusted covariates (age, stage, performance status, and response to ASCT). In treatment benefit analysis, PFS and OS were modeled with genotype, treatment assignment, and their interaction term. Logistic regression models were used for analysis of thalidomide-related peripheral neuropathy with time to worsening of neuropathy by at least 1 grade using the National Cancer Institute Common Toxicity Criteria (NCI-CTC version 2.0)21 being the outcome of interest. The dominant genetic model was used for genotype grouping (similar results with the additive model). A 2-sided P value of <.05 was required for statistical significance. The Research Ethics Board of University of New Brunswick, Canada provided institutional review board approval for this study (REB#03-2013).
We identified SNPs with significant prognostic impact on PFS and/or OS (Table 1). In a multivariate analysis with inclusion of known prognostic factors, all SNPs retained significant associations (supplemental Table 3). Five SNPs had significant interaction with treatment allocation in predicting PFS and/or OS (Table 2). Two SNPs were found to have significant interaction with treatment allocation in predicting time to worsening of peripheral neuropathy (Table 3).
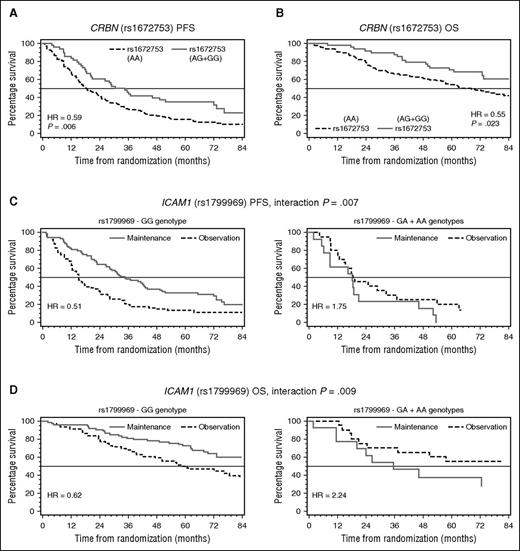
A SNP (rs1672753) in the 5′ untranslated region (UTR) of CRBN had previously been associated with immunomodulatory drug (IMiD) response in myelodysplastic patients.12 In the MY.10 cohort, this SNP had significant impact on myeloma survival outcome independent of thalidomide maintenance therapy. Patients with GG and GA genotypes had better PFS and OS compared with patients with 2 major alleles (AA) (Figure 1A-B). Although CRBN has been shown to play a central role in the mechanism of action of IMiDs,22 the prognostic impact of this SNP regardless of treatment arm suggests that CRBN may play a role in myeloma cell biology that is independent of IMiD therapy.
Kaplan-Meier plot comparing survival of patients from the MY.10 clinical trial by SNP status. HR of CRBN (rs1672753) represent hazard of patients with at least 1 minor allele vs patients with 2 major alleles. HR of ICAM1 (rs1799969) represents hazard of treatment (maintenance) vs observation for each genotype group.
Kaplan-Meier plot comparing survival of patients from the MY.10 clinical trial by SNP status. HR of CRBN (rs1672753) represent hazard of patients with at least 1 minor allele vs patients with 2 major alleles. HR of ICAM1 (rs1799969) represents hazard of treatment (maintenance) vs observation for each genotype group.
SNPs in cytochrome P450 genes found to be prognostic for PFS and/or OS in our cohort (CYP1A1 rs4646903 and CYP1B1 rs1056836) may in fact be associated with melphalan benefit. The cytochrome P450 family of enzymes is known to mediate chemotherapeutic drug metabolism and activation.23 We were unable to correlate these SNPs directly with melphalan benefit because all patients received melphalan as a conditioning agent. These SNPs were found previously by Dumontet et al to be associated with outcomes after high-dose melphalan,8 with the same direction of association.
Patients with AA and AG genotypes for SNP rs1799969 (ICAM1) had inferior hazard ratio (HR) estimates for PFS and OS on maintenance therapy compared with observation, suggesting possible harm from maintenance therapy; conversely, patients with 2 major alleles (GG) had superior HR estimates for PFS and OS on maintenance compared with observation, suggesting benefit from maintenance therapy (Figure 1C-D). This SNP was found previously to be associated with decreased risk of thalidomide-related peripheral neuropathy7 ; we did not detect such an association in our study. ICAM1 is an intracellular adhesion molecule involved in myeloma cell adhesion-mediated drug resistance (CAMDR). Minor (C) allele correlates with low ICAM1 levels.24 Thalidomide is able to downregulate ICAM1.25 High ICAM1 levels would normally lead to CAMDR and melphalan resistance. Patients with 2 major alleles in the observation group would be melphalan resistant, leading to poorer survival outcome. If these patients receive subsequent thalidomide maintenance therapy and overcome CAMDR, this could result in an improved survival outcome compared with observation.
GLCCI1 SNP (rs37973) assayed in this study is always coinherited with SNP rs37972, which has previously been associated with corticosteroid response in asthma patients.19 Because MY.10 patients in the maintenance arm received prednisone, it is intriguing to speculate on the potential modulation of neuropathy by corticosteroid therapy.
Due to the limited sample size and exploratory nature of this study, correction for multiple hypothesis testing was not conducted, and raises the potential for both false-negative and false-positive results. Firm conclusions cannot be drawn regarding SNPs that were significant in previous studies but were not validated in our study, and SNPs identified as significant for the first time in our study require independent validation in future patient cohorts. Functional studies will help clarify our findings.
This is the first randomized trial with a no-treatment control arm to report the impact of SNPs in predicting benefit from immunomodulatory drug maintenance therapy for myeloma. Thalidomide biomarkers discovered here may apply to lenalidomide and pomalidomide, due to their structural similarity and common mechanisms of action.
Presented in abstract form at the 57th annual meeting of the American Society of Hematology, Orlando, FL, December 5-8, 2015.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank Bithika Ray for procuring reagents and supplies, and Wei Xu for developing the initial plan for statistical analysis.
This work was supported by a grant from Canada’s Foundation for Innovation (T.R.). M.H. is supported by a trainee award from the Beatrice Hunter Cancer Research Institute with funds provided by Dalhousie Medicine New Brunswick and Cancer Care Nova Scotia as part of The Terry Fox Strategic Health Research Training Program in Cancer Research at the Canadian Institutes of Health Research (CIHR).
M.H. is a candidate at University of New Brunswick, and this work is submitted in partial fulfillment of the requirement for the MSc degree. T.R. is the Canadian Cancer Society Research Chair at the University of New Brunswick.
Contribution: M.H. performed experiments and wrote the paper; B.E.C. developed the statistical analysis plan and analyzed results; M.D. coordinated decoding of patient identifiers; A.M. and T.R. designed the research; A.M., T.R., and Y.X. helped edit the manuscript; N.J.B., K. Song, D.W., C.C., M.D.S., K.H.-J., D.R., K. Stewart, R.M.M., L.Z., and T.R. recruited patients to the MY.10 clinical trial and collected patient samples for this study; K. Stewart, A.E.H., M.D., R.M.M., and L.S. were involved in the original study design, review, and analysis of the data and facilitated the patient sample collection.
Conflict-of-interest disclosure: N.J.B. has consulted for Celgene and Amgen, and received research funding from Johnson & Johnson. K. Stewart. is a member of the advisory committee and received honoraria and research funding from Celgene Canada. D.W., C.C., M.D.S., L.S., B.E.C., M.D., K.H.-J., and T.R. received honoraria and research funding from Celgene Canada. D.R. is a consultant at Celgene, Janssen-Cilag, and Onyx, and received honoraria and research funding from Celgene, Janssen-Cilag, Onyx, Amgen, Novartis, Lundbeck, Merck, Bristol-Myers Squibb, Otsuka, and Millennium Takeda. The remaining authors declare no competing financial interests.
Correspondence: Tony Reiman, Department of Oncology, Saint John Regional Hospital, 400 University Ave, PO Box 2100, Saint John, NB E2L4L2, Canada; e-mail: anthony.reiman@horizonnb.ca.