Key Points
OS of patients with FLT3-ITD differs significantly when categorized by the 2017 ELN risk stratification.
In a multivariate Cox model for OS, there is a consistent beneficial effect of midostaurin across the 3 2017 ELN risk groups.
Abstract
Patients with acute myeloid leukemia (AML) harboring FLT3 internal tandem duplications (ITDs) have poor outcomes, in particular AML with a high (≥0.5) mutant/wild-type allelic ratio (AR). The 2017 European LeukemiaNet (ELN) recommendations defined 4 distinct FLT3-ITD genotypes based on the ITD AR and the NPM1 mutational status. In this retrospective exploratory study, we investigated the prognostic and predictive impact of the NPM1/FLT3-ITD genotypes categorized according to the 2017 ELN risk groups in patients randomized within the RATIFY trial, which evaluated the addition of midostaurin to standard chemotherapy. The 4 NPM1/FLT3-ITD genotypes differed significantly with regard to clinical and concurrent genetic features. Complete ELN risk categorization could be done in 318 of 549 trial patients with FLT3-ITD AML. Significant factors for response after 1 or 2 induction cycles were ELN risk group and white blood cell (WBC) counts; treatment with midostaurin had no influence. Overall survival (OS) differed significantly among ELN risk groups, with estimated 5-year OS probabilities of 0.63, 0.43, and 0.33 for favorable-, intermediate-, and adverse-risk groups, respectively (P < .001). A multivariate Cox model for OS using allogeneic hematopoietic cell transplantation (HCT) in first complete remission as a time-dependent variable revealed treatment with midostaurin, allogeneic HCT, ELN favorable-risk group, and lower WBC counts as significant favorable factors. In this model, there was a consistent beneficial effect of midostaurin across ELN risk groups.
Introduction
Activating mutations of FLT3 are among the most common mutations in patients with acute myeloid leukemia (AML).1-3 There are 2 major types of mutations, internal tandem duplications (ITDs), and mutations within the activation loop of the second tyrosine kinase domain.4 ITDs are in-frame duplications that involve different functional domains of the receptor, most commonly the juxtamembrane domain, and lead to constitutive activation of the receptor.4 FLT3-ITD has consistently been associated with higher white blood cell (WBC) counts, higher percentages of bone marrow (BM) blast cells, an increased risk for relapse, and inferior survival.5-7
Factors that have been shown to influence the prognostic impact of FLT3-ITDs are the mutational context, in particular the mutational status of NPM1,8-10 the insertion site of the ITD,11-13 and, importantly, the allelic ratio (AR),14-16 which is most commonly assessed by DNA fragment analysis using a polymerase chain reaction (PCR)-based method combined with capillary electrophoresis. Recent studies have indicated that patients with NPM1 mutation (NPM1mut) and concurrent FLT3-ITD with a low (<0.5) AR (FLT3-ITDlow) have a favorable outcome that is similar to patients with NPM1mut and wild-type FLT3 (FLT3wt).8,9,15,16 In contrast, patients with wild-type NPM1 (NPM1wt) and FLT3-ITD with a high (≥0.5) AR (FLT3-ITDhigh) have a poor outcome.9,17
Another important observation relates to the impact of allogeneic hematopoietic cell transplantation (HCT) in patients with these different genotypes. Several groups have shown that patients with the more favorable genotype NPM1mut/FLT3-ITDlow may not derive benefit from allogeneic HCT as first-line treatment,10,15,18 although this effect has not been observed in all studies.19,20 The preponderant evidence for the prognostic significance of the FLT3-ITD AR is now reflected in the 2017 European LeukemiaNet (ELN) recommendations that distinguish prognostic NPM1/FLT3-ITD genotypes by accounting for the FLT3-ITD AR.21
The natural course of AML with FLT3 mutation may change with the use of FLT3 inhibitors, and the prognostic impact of the above genotypes will need to be revisited.22 Midostaurin, a multikinase inhibitor, is a first-generation FLT3 inhibitor.23 Based on the results of the international randomized CALGB 10603/RATIFY study, midostaurin was approved, in combination with intensive chemotherapy, by the US Food and Drug Administration and by the European Medicines Agency; in addition, it was approved as maintenance therapy for adult patients with AML exhibiting an activating FLT3 mutation by the European Medicines Agency.24 Further evidence for the efficacy of midostaurin in patients with FLT3-ITD+ AML comes from the AMLSG 16-10 trial, which also included older patients aged 60 to 70 years.25
The objectives of this study were to validate the prognostic impact of the NPM1/FLT3-ITD genotypes, as defined by the 2017 ELN recommendations, and to evaluate the potential predictive impact of these genotypes for response to midostaurin in randomized patients from the RATIFY trial.
Patients and methods
Patients
Overall, 717 patients with AML and activating FLT3 mutations (ITD and tyrosine kinase domain mutations) were included in the CALGB 10603/RATIFY trial.24 This post hoc exploratory analysis focuses on the subset of patients with FLT3-ITD.
Data on the 4 NPM1/FLT3-ITD genotypes, considering the FLT3-ITD AR (low, 0.05 to <0.5; high, ≥ 0.5), were available in 427 of 549 patients with FLT3-ITD AML. Table 1 shows the characteristics of these patients. The 2017 ELN high-risk markers RUNX1, ASXL1, and TP53 could be assessed in 358 of these patients who gave informed consent for further molecular studies and for whom DNA was still available. Table 1 shows how these high-risk markers segregated among the 4 NPM1/FLT3-ITD genotypes. The study was approved by the Institutional Review Board of Ulm University.
Complete 2017 ELN risk categorization could be done for 318 of 549 patients. Table 2 provides the clinical and genetic characteristics of these 318 patients by risk group. Baseline characteristics between the clinical trial cohort of all FLT3-ITD+ patients (n = 549) and the ELN biomarker cohort (n = 318) were balanced, as were complete remission (CR) rates and overall survival (OS) times (supplemental Table 1; supplemental Figure 1, available on the Blood Web site).
Genetic analyses
FLT3-ITD mutation analysis was performed as described.24 Testing was done in 9 reference laboratories in 6 countries using a harmonized PCR method based on capillary electrophoresis detection. PCR was done in triplicate, and the mean values of these measurements were reported. To ensure consistency among laboratories, a cross-validation quality control procedure was performed every 6 months.26 Patients were eligible for the clinical trial when exceeding the diagnostic cutoff for the FLT3-ITD AR (≥0.05). Randomization to midostaurin vs placebo was stratified by the ITD AR (low, 0.05-0.7; high, >0.7).6 For this analysis, we chose a cutoff for the AR of 0.5 (low, >0.05 to <0.5; high, ≥0.5), because this cutoff value has been shown to better discriminate and has also been adopted in the 2017 ELN risk classification.10,15,21 Semiquantitative assessment of FLT3-ITD AR (using DNA fragment analysis) was determined as the area under the curve “FLT3-ITD” divided by the area under the curve “FLT3wt.”21
Statistical analysis
CR was defined by standard criteria21 ; responses included all CRs achieved during induction cycles 1 and 2. The definition of OS, event-free survival (EFS), cumulative incidence of relapse (CIR), and cumulative incidence of death (CID) were based on recommended criteria.21 Survival times were calculated from the date of randomization. The median follow-up for survival was calculated using the reverse Kaplan-Meier estimate.29 Logistic regression and Cox proportional hazards models were used to identify prognostic variables for CR, OS, and CIR.30 Additional covariates in multivariate analysis were age, BM blast counts, and WBC counts as continuous variables and sex and treatment (midostaurin vs placebo) as dichotomous variables; allogeneic HCT was included as a time-dependent variable. The subgroup results of proportional hazards models were summarized in forest plots. Comparisons between the NPM1/FLT3-ITD genotypes and the 2017 ELN risk groups with respect to quantitative variables were performed using the Kruskal-Wallis test. Survival distributions were estimated using the Kaplan-Meier method,31 and differences between groups were analyzed using 2-sided log-rank tests.
To estimate survival probabilities considering the effect of allogeneic HCT in first CR (CR1), the Simon-Makuch method was used with clock-back correction, according to Bernasconi et al.32 Simon-Makuch estimates show the survival probabilities for fictional patients who either never receive an allogeneic HCT or have received an allogeneic HCT at t = 0. To examine the effect of allogeneic HCT, univariate and multivariate Cox models, with allogeneic HCT as a time-dependent intervening event, were applied.33 An effect was considered significant if its P value was <5%. The analyses were not adjusted for multiple testing.
Leave-1-out cross-validated prediction errors were used to evaluate the prognostic value of the 2017 ELN risk classification, in which the prediction error is defined using Brier’s score as a function of time.34 To account for allogeneic HCT in CR1 as a time-dependent intervention, prediction errors were calculated using multistate models.35 The “reference” model is the Aalen-Johansen estimator.36 For ordinary (single-event) survival this reduces to the Kaplan-Meier estimate. All statistical analyses were performed with statistical software (R 3.5.1).
Results
Categorization of patients
The 427 patients, for whom data on FLT3-ITD AR and NPM1 mutational status were available, were first categorized to 1 of the 4 NPM1/FLT3-ITD genotypes: NPM1mut/FLT3-ITDlow (n = 85, 19.9%), NPM1mut/FLT3-ITDhigh (n = 159, 37.2%), NPM1wt/FLT3-ITDlow (n = 74, 17.4%), and NPM1wt/FLT3-ITDhigh (n = 109, 25.5%). Patient and disease characteristics according to these genotypes are given in Table 1. Patients with concurrent NPM1mut were older and more frequently female; patients with high FLT3-ITD AR had higher WBC counts and higher BM blast counts. Patients with NPM1mut AML more frequently had a normal karyotype compared with patients with NPM1wt AML. With regard to the concurrent presence of 2017 ELN high-risk markers, NPM1mut was almost mutually exclusive with RUNX1 mutations, whereas 30.2% and 26.1% of the NPM1wt/FLT3-ITDlow and NPM1wt/FLT3-ITDhigh genotypes, respectively, had RUNX1 mutations; ASXL1 mutations were distributed more equally, and TP53 mutations were only found in 2 cases.
We subsequently categorized patients according to the 2017 ELN risk groups (supplemental Table 2). Complete categorization could be done for 318 patients (“ELN biomarker cohort”): (1) favorable risk (n = 85, ie, NPM1mut/FLT3-ITDlow AML), (2) intermediate risk (n = 111, ie, NPM1mut/FLT3-ITDhigh AML [n = 93] and NPM1wt/FLT3-ITDlow AML [n = 18], both subgroups without the concurrent presence of the high-risk molecular markers RUNX1, ASXL1, TP53, as well as NPM1wt/FLT3-ITDlow AML without adverse-risk cytogenetics; and (3) adverse risk (n = 122, NPM1wt/FLT3-ITDhigh AML [n = 92], NPM1mut/FLT3-ITDhigh AML [n = 8] exhibiting high-risk molecular markers, and NPM1wt/FLT3-ITDlow AML [n = 22] with high-risk molecular markers and/or adverse-risk cytogenetics). Patient and disease characteristics are given in Table 2.
Response to induction therapy
We assessed response to therapy by ELN risk group and by treatment arm (midostaurin vs placebo). Responses included all CRs achieved during induction cycles 1 and 2 (Table 2). Responses in patients with favorable, intermediate, and adverse risk were as follows: with placebo, 68.1% vs 59.6% vs 44.4%, respectively (P = .05); with midostaurin, 71.1% vs 66.7% vs 57.4%, respectively (P = .34). There was no significant difference in response between treatment arms in the 3 ELN risk groups.
In multivariable logistic regression analysis, factors for lower CR rates were ELN adverse vs favorable risk (odds ratio [OR], 0.54; 95% confidence interval [CI], 0.29-0.99; P = .052) and higher WBC (10-fold) (OR, 0.62; 95% CI, 0.39-0.97; P = .039). Age (difference of 10 years; OR, 1.07, 95% CI, 0.85-1.34; P = .55), sex (female vs male; OR, 1.00; 95% CI, 0.62-1.63; P = .99), treatment (midostaurin vs placebo; OR, 1.26; 95% CI, 0.78-2.03; P = .35), and BM blasts (twofold) (OR, 0.96; 95% CI, 0.66-1.38; P = .84) did not have a significant influence.
Outcomes
The estimated median follow-up of the 318 patients was 57.5 months (95% CI, 55.2-61.2). Median OS and 5-year OS rate were 26.3 months (95% CI, 18.6-50.7) and 0.44 (95% CI, 0.39-0.50); median EFS and 5-year EFS rate were 4.70 months (95% CI, 1.97-7.49) and 0.24 (95% CI, 0.20-0.29). Median OS and 5-year OS rates in the placebo and midostaurin arms were 16.6 months (95% CI, 13.9-23.2) and 0.34 (95% CI, 0.27-0.43) and not reached (95% CI, 29.8 months-not reached) and 0.53 (95% CI, 0.46-0.61), respectively.
Survival analysis by ELN risk groups
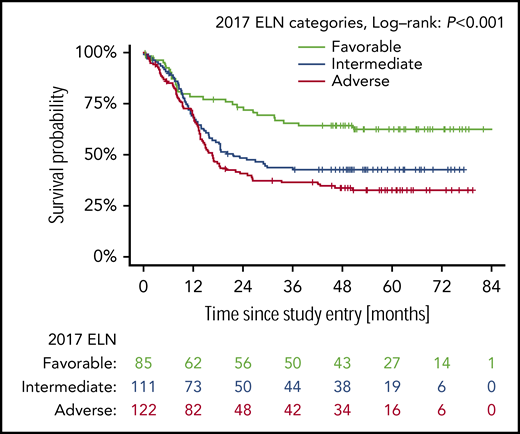
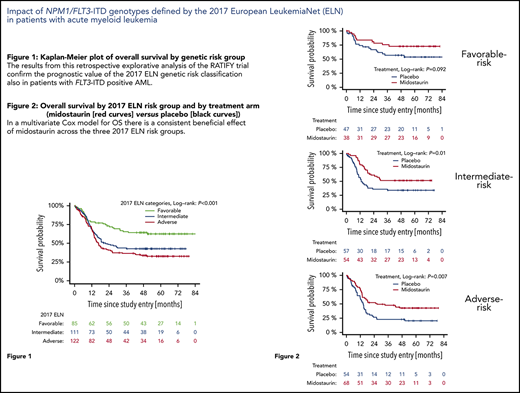
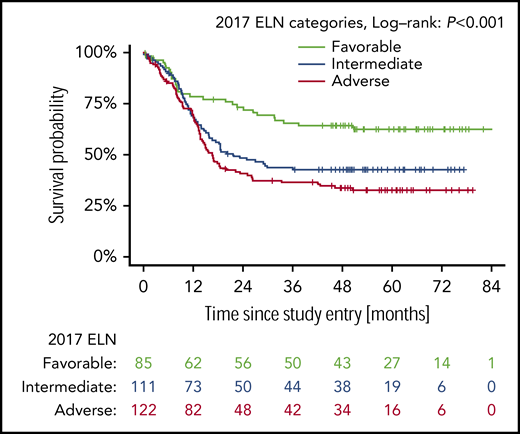
Figure 1 shows OS according to the 2017 ELN risk groups. Patients with favorable-risk AML had the best outcome (5-year OS probability 0.62; 95% CI, 0.53-0.74), followed by patients with intermediate-risk AML (0.43; 95% CI, 0.34-0.53) and patients with adverse-risk AML (0.33; 95% CI, 0.25-0.42). The difference between the curves was statistically significant (P < .001). Supplemental Figure 2A shows the prediction error curve for the 2017 ELN risk classification in comparison with the marginal Kaplan-Meier reference. An advantage of the ELN 2017 risk classifier is observed from 1-year follow-up onward.
Prognostic effect on overall survival of patients with the different NPM1/FLT3-ITD genotypes categorized according to 2017 ELN genetic risk groups. The P values for the log-rank tests comparing favorable vs intermediate and intermediate vs adverse are P = .007 and P = .20, respectively.
Prognostic effect on overall survival of patients with the different NPM1/FLT3-ITD genotypes categorized according to 2017 ELN genetic risk groups. The P values for the log-rank tests comparing favorable vs intermediate and intermediate vs adverse are P = .007 and P = .20, respectively.
For illustration purposes, we included supplemental Figure 3, which shows OS according to the 4 NPM1/FLT3-ITD genotypes not categorized according to the ELN risk groups. Supplemental Figure 4 shows the forest plot of hazard ratios (HRs) from the treatment arm (midostaurin vs placebo), derived from univariate Cox models, by NPM1/FLT3-ITD genotypes.
Outcome analysis by ELN risk group and by treatment arm
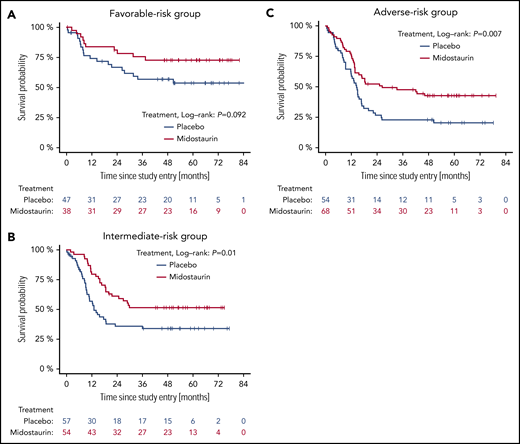
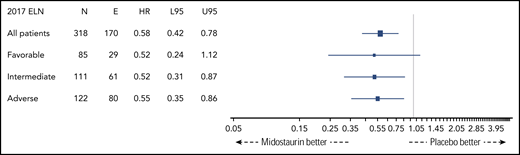
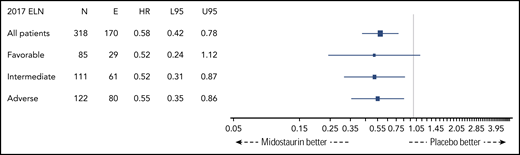
Figure 2 shows OS by ELN risk group and by treatment arm (midostaurin vs placebo). Five-year OS rates of patients on the midostaurin and the placebo arm were 0.73 (95% CI, 0.60-0.89) and 0.53 (95% CI, 0.40-0.72), 0.52 (95% CI, 0.40-0.67) and 0.34 (95% CI, 0.23-0.49), and 0.43 (95% CI, 0.32-0.56) and 0.20 (95% CI, 0.12-0.35) in the favorable-, intermediate-, and adverse-risk groups, respectively. The corresponding forest plot of HRs for OS is shown in Figure 3. A beneficial effect of midostaurin was observed across all 3 risk groups. No treatment effect modification by the 2017 ELN risk classification was found.
OS of patients with the different NPM1/FLT3-ITD genotypes by 2017 ELN risk group and by treatment. (A) Favorable-risk group. (B). Intermediate-risk group. (C) Adverse-risk group.
OS of patients with the different NPM1/FLT3-ITD genotypes by 2017 ELN risk group and by treatment. (A) Favorable-risk group. (B). Intermediate-risk group. (C) Adverse-risk group.
Forest plot of HRs of treatment arm (midostaurin vs placebo) derived from univariate Cox models by 2017 ELN risk groups. E, number of events; L95, lower 95% CI; N, number of patients; U95, upper 95% CI.
Forest plot of HRs of treatment arm (midostaurin vs placebo) derived from univariate Cox models by 2017 ELN risk groups. E, number of events; L95, lower 95% CI; N, number of patients; U95, upper 95% CI.
A multivariate Cox model for OS of the entire cohort, including the covariates ELN risk groups, age, WBC count, BM blasts, sex, allogeneic HCT in CR1, and treatment, identified the 2017 ELN risk classification, treatment (midostaurin vs placebo), allogeneic HCT in CR1, and log10 WBC count as significant prognostic variables (Table 3). Leave-1-out cross-validated prediction error curves, according to Spitoni et al,35 demonstrate the added value of the 2017 ELN classifier for 1-year follow-up and beyond, comparing the models with and without the 2017 ELN risk classification (supplemental Figure 2B).
CIR by ELN risk group, as well as by ELN risk group and by treatment arm, is shown in supplemental Figures 5A and 6, respectively. Five-year CIR rates for patients on the midostaurin and the placebo arms were 0.22 (95% CI, 0.07-0.38) and 0.35 (95% CI, 0.19-0.52), 0.36 (95% CI, 0.20-0.52) and 0.62 (95% CI, 0.45-0.78), and 0.58 (95% CI, 0.42-0.74) and 0.61 (95% CI, 0.41-0.81) in the favorable-, intermediate-, and adverse-risk groups, respectively. Supplemental Figures 5B and 7 show CID by ELN risk group and by treatment arm. In the adverse-risk group, there was a significantly lower CID rate with midostaurin (0.05; 95% CI, 0-0.12) vs placebo (0.26; 95% CI, 0.08-0.44; P = .024), providing an explanation for the discrepancy between improved OS on the midostaurin arm without decreasing the relapse rate. A multivariate Cox model for CIR identified ELN risk classification, treatment with midostaurin, allogeneic HCT in CR1, and WBC count as independent prognostic factors (supplemental Table 3). Supplemental Figure 8 displays the leave-1-out cross-validated prediction error curves according to Spitoni et al,35 indicating the advantage of the 2017 ELN classifier, comparing the models with and without the 2017 ELN risk classification.
EFS by ELN risk group and treatment arm is shown in supplemental Figure 9.
Impact of allogeneic HCT in the ELN risk groups
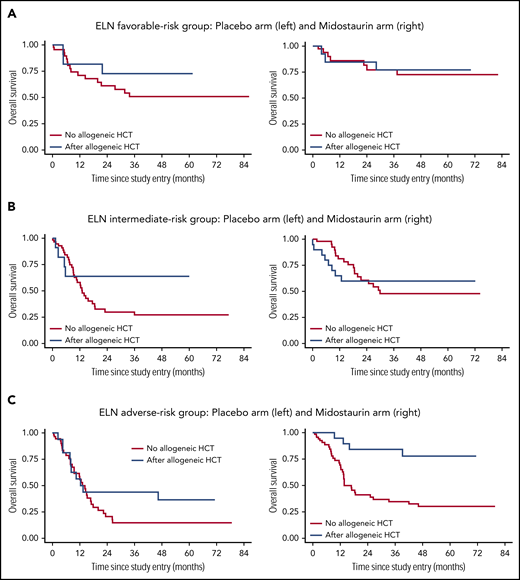
To illustrate the effect of allogeneic HCT in CR1 on OS, the Simon-Makuch method was used to estimate survival probabilities with respect to time-dependent interventions.32 Of note, there was a consistent beneficial effect of midostaurin across all 3 risk groups, whereas this was not evident for allogeneic HCT. A strong beneficial effect of allogeneic HCT was only found in the adverse-risk group, with an HR of 0.39 (95% CI; 0.21-0.73; P = .003). Table 3 summarizes results from the multivariate Cox model for OS in the entire cohort, as well as in the 3 ELN risk groups. Figure 4 provides Simon-Makuch plots illustrating OS by 2017 ELN risk group, type of postremission therapy (conventional consolidation vs allogeneic HCT in CR1), and treatment arm (placebo vs midostaurin). Supplemental Figure 10 shows the corresponding Simon-Makuch plots by 2017 ELN risk group and by type of postremission therapy only.
OS by 2017 ELN risk group, by type of postremission therapy (conventional consolidation vs allogeneic HCT in CR1), and by treatment arm (placebo vs midostaurin). Simon-Makuch plots illustrating the influence of allogeneic HCT as a time-dependent variable: patients who receive an allogeneic HCT move from the red to the blue curve at the time that the allogeneic HCT is performed. (A) ELN favorable-risk group: placebo arm (left panel) and midostaurin arm (right panel). (B) ELN intermediate-risk group: placebo arm (left panel) and midostaurin arm (right panel). (C) ELN adverse-risk group: placebo arm (left panel) and midostaurin arm (right panel).
OS by 2017 ELN risk group, by type of postremission therapy (conventional consolidation vs allogeneic HCT in CR1), and by treatment arm (placebo vs midostaurin). Simon-Makuch plots illustrating the influence of allogeneic HCT as a time-dependent variable: patients who receive an allogeneic HCT move from the red to the blue curve at the time that the allogeneic HCT is performed. (A) ELN favorable-risk group: placebo arm (left panel) and midostaurin arm (right panel). (B) ELN intermediate-risk group: placebo arm (left panel) and midostaurin arm (right panel). (C) ELN adverse-risk group: placebo arm (left panel) and midostaurin arm (right panel).
Discussion
The results from this retrospective explorative analysis of the RATIFY trial confirm the prognostic value of the 2017 ELN genetic risk classification in patients with FLT3-ITD+ AML. Furthermore, the data show that midostaurin exerts a beneficial effect in these patients across all 3 ELN risk groups.
Randomization in the RATIFY trial was stratified by the FLT3-ITD AR with a cutoff > 0.7 vs 0.05 to 0.7.24 This cutoff was chosen based on the initial study reported by Thiede et al.6 More recent studies found an ITD AR of 0.5 to be a better discriminator for prognosis.10,15 Based on these more recent studies, the 2017 ELN recommendations put forward the current NPM1/FLT3-ITD risk categories that are based on an ITD AR cutoff of 0.5 and the NPM1 mutational status.21 An important aspect relates to the diagnostic assay for assessment of the AR. In the RATIFY trial, testing for FLT3-ITD was done in 9 reference laboratories in 6 countries using a harmonized PCR method based on capillary electrophoresis detection. To ensure a high degree of consistency among laboratories, a cross-validation quality control procedure was performed every 6 months. The assessment of the ITD AR showed variability, which could be reduced by using a triplicate analysis.26 Nevertheless, similar to many other diagnostic assays, there is a need for further harmonization and standardization of the testing.
The 4 NPM1/FLT3-ITD genotypes were associated with significant differences in clinical and concurrent genetic features. As previously shown,27 compared with patients with NPM1wt, more FLT3-ITD patients with concurrent NPM1mut were female and had a normal karyotype. Patients with a high FLT3-ITD allelic burden had significantly higher WBC counts and higher BM blast numbers. The genotypes also differed significantly with regard to the concurrent presence of the 2017 ELN high-risk molecular markers RUNX1, ASXL1, and TP53. RUNX1 mutations were almost mutually exclusive of NPM1mut. The highest frequencies of RUNX1 mutations were found in NPM1wt/FLT3-ITDlow AML (30.2%), moving these cases from the intermediate-risk group to the adverse-risk group, as well as in NPM1wt/FLT3-ITDhigh AML (26.1%). ASXL1 mutations were distributed more equally among the 4 NPM1/FLT3-ITD genotypes, and TP53 mutations were only found in 2 AML patients.
Complete categorization of the NPM1/FLT3-ITD genotypes according to the 2017 ELN classification could be done for 318 of the 549 trial patients with FLT3-ITD+ AML. The first important finding of this study is that the data confirm the high prognostic value of the 2017 ELN risk categorization, also among patients with FLT3-ITD+ AML (Figure 1). In particular, the data provide further evidence for the favorable prognosis of patients with NPM1mut/FLT3-ITDlow AML. Five-year OS for patients in the 2 treatment arms combined was 62.5%; it was 73.0% in patients treated on the midostaurin arm, which is comparable to the outcome of patients with the other more favorable-risk AML, such as core-binding factor AML37,38 and AML with biallelic CEBPA mutations.39 Five-year OS for patients in the ELN adverse-risk group was 32.7%; it was 43.0% on the midostaurin arm. Compared with historical controls, outcomes for the adverse-risk patient group appear to be significantly improved by the addition of midostaurin and by allogeneic HCT.
The second important finding is that midostaurin had a beneficial effect on OS in FLT3-ITD AML across all 3 ELN risk groups (Figures 2 and 3). Thus, midostaurin appears to be active in FLT3-ITD+ AML, irrespective of the allelic burden of the ITD, and, importantly, as well as on different mutational backgrounds (eg, with or without NPM1mut) and with or without selected adverse-risk genetic features. The effect on CIR appeared to be most pronounced in the ELN intermediate-risk group (supplemental Figure 6). The fact that midostaurin has a beneficial effect on OS, independent of the ITD allelic burden and across various underlying genetic signatures, raises the question of whether the therapeutic effect is primarily mediated through its FLT3-inhibitory effect or through other antileukemic effects of this multikinase inhibitor.40
Finally, an important point of discussion in the past has been the value of allogeneic HCT in patients with the different NPM1/FLT3-ITD genotypes.10,15,17-19 To address the impact of allogeneic HCT in CR1, we used the Simon-Makuch method to estimate survival distributions of time-dependent interventions.32 Multivariate analysis for OS using the Mantel-Byar test in the entire patient cohort identified treatment with midostaurin, allogeneic HCT, 2017 ELN favorable risk, and lower WBC counts as significant favorable factors for OS (Table 3). The same variables were identified in the multivariate model for cause-specific hazard of relapse (supplemental Table 3). Next, we performed multivariate analysis for OS within the 3 ELN risk groups. Of note, the only variable that showed a consistent favorable effect across all risk groups was treatment with midostaurin. For allogeneic HCT, in contrast, a strong beneficial effect was only observed in the adverse-risk group. Figure 4 provides Simon-Makuch plots illustrating the influence of allogeneic HCT vs conventional consolidation on OS in the 3 risk groups and by treatment arm (placebo vs midostaurin). These results need to be interpreted with caution because the clinical trial was not powered to show statistically significant differences in these genetic subgroups with regard to allogeneic HCT and with regard to treatment with midostaurin vs placebo. Nevertheless, the data provide further evidence that conventional consolidation plus midostaurin is a postremission treatment option for patients with FLT3-ITD, ELN favorable-risk AML, and allogeneic HCT may be delayed until first relapse in this patient population. Importantly, in these patients, NPM1mut provides a solid target for the monitoring of measurable residual disease, allowing for further refinement of the prognostic assessment, which taken all together, will inform the most appropriate postremission therapy.41,42 Based on the results of this study, one could also envision a similar treatment strategy for many patients with FLT3-ITD, ELN intermediate-risk AML, the majority of whom also carry concurrent NPM1mut. This treatment algorithm should be explored in future studies.
Of note, the RATIFY trial only recruited patients 18 to 60 years of age; thus, the data provided by this retrospective exploratory study cannot be automatically extrapolated to patients older than 60 years. Nevertheless, the AMLSG 16-10 trial, evaluating midostaurin in patients 18 to 70 years of age, reported very encouraging results in patients 60 to 70 years of age, as well.25
In conclusion, the data from this study provide further support for the high prognostic value of the 2017 ELN risk categorization in patients with FLT3-ITD. A complete work-up according to 2017 ELN recommendations, including assessment of the ITD allelic burden, should be mandatory for all newly diagnosed FLT3-ITD patients eligible for intensive therapy. The multikinase inhibitor midostaurin showed a beneficial effect across all risk groups and independent of allogeneic HCT. This study further stresses the beneficial impact of allogeneic HCT in patients with FLT3-ITD, ELN adverse-risk AML.
Presented in part in abstract form at the 59th annual meeting of the American Society of Hematology, Atlanta, GA, 7 December 2017.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (SFB 1074, project B3) (K.D.), by National Institutes of Health, National Cancer Institute grants U10CA180821 (to the Alliance for Clinical Trials in Oncology Operations Center), U10CA180882 (to the Alliance Statistics and Data Management Center), U24CA196171 (to the Alliance NCTN Biorepository and Biospecimen [C.D.B.]), and UG1CA233338 (to ITSC for Leukemia: Novel Molecular strategies for NCTN Individualized Therapies [C.D.B.]), and by a research grant from Novartis.
Authorship
Contribution: K.D. and C.T. designed the study, performed research, collected, assembled, analyzed, and interpreted data, and wrote the manuscript; N.J., E.P., T.W.P., D.J., M.H., T.O., J.F.N., and J.H.J. performed molecular analyses and analyzed data; A. Gambietz, S.J.M., I.G., and A.B. performed statistical analyses; R.A.L., G.M., R.B.K., A.H.W., J.S., M.A.S., J.M.B., T.d.W., D.N., F.R.A., B.C.M., M.S.T., R.F.S., A. Ganser, H.S., G.E., S.A., and C.P. collected, assembled, analyzed, and interpreted data; J.K. and M.T.V. collected and assembled data, performed molecular analyses, and interpreted and analyzed data; and R.M.S., H.D., and C.D.B. designed the study, collected, assembled, analyzed and interpreted data, and wrote the manuscript. All authors approved the final version of the manuscript.
Conflict-of-interest disclosure: K.D. has acted as a consultant or advisor for Astellas, Celgene, Daiichi Sankyo, Janssen, Novartis, and Roche and has received clinical research support from Astex, Celgene, and Novartis. C.T. is the Chief Executive Officer and a co-owner of Agendix, a company performing molecular diagnostics; has acted as a consult or advisor for Novartis and Astellas; and has received clinical research support from Bayer. R.A.L. has acted as a consultant or advisor and has received clinical research support from Novartis. G.M., J.F.N., R.B.K., D.N., and H.S. have acted as consultants or advisors for Novartis. D.J. and A. Ganser have received clinical research support from Novartis. J.K. has acted as a consultant or advisor for Amgen, Astellas, Bristol-Myers Squibb, Daiichi Sankyo, and Pfizer. M.H. has acted as a consultant or advisor for AbbVie, Daiichi Sankyo, Novartis, Pfizer, and Bayer Pharma and has received clinical research support from Pfizer, Daiichi Sankyo, Karyopharm, BerGenBio, Bayer Pharma, Novartis, and Astellas. S.J.M. has acted as a consultant or advisor for Pfizer and Pique Therapeutics; has other relationship with BeiGene. A.H.W. has acted as a consultant or advisor for Novartis, Astellas, Pfizer, MacroGenics, AbbVie, Genentech, Servier, Celgene, Amgen, Astra Zeneca, and Janssen; is a member of the speakers bureau for AbbVie/Genentech and Novartis; has received research funding from Novartis, Celgene, AbbVie, Servier, Astra Zeneca, and Amgen; and is a former employee of the Walter and Eliza Hall Institute and receives a fraction of its royalty stream related to venetoclax. J.S. has acted as a consultant or advisor for Pfizer, Daiichi Sankyo, AbbVie, Novartis, Astellas, and Roche and is a member of the speakers bureau for Novartis, Pfizer, Daiichi Sankyo, and AbbVie. M.A.S. has acted as a consultant or advisor for Teva Pharmaceutical Industries, Daiichi-Sankyo, Orsenix, AbbVie, Novartis, and Pfizer. T.d.W. has acted as a consultant or advisor for Novartis, Celgene, Johnson & Johnson, and Incyte and has received clinical research support from Novartis, Celgene, and Johnson & Johnson. B.C.M. has acted as a consultant or advisor for Celgene, Novartis, and Astellas and is currently employed by Roche/Genentech. M.T. has acted as a consultant or advisor for AbbVie, BioLineRx, Daiichi-Sankyo, Orsenix, KAHR Medical, Rigel Pharmaceuticals, Nohla, Delta Fly Pharma, Tetraphase, Oncolyze, and Jazz Pharmaceuticals; has received clinical research funding from AbbVie, Cellerant Therapeutics, Orsenix, ADC Therapeutics, and BioSight; and has received royalties from UpToDate. R.F.S. has acted as a consultant or advisor for Pfizer and Daiichi Sankyo; is a member of the speakers bureau for Novartis, Pfizer, and Daiichi Sankyo; and has received clinical research funding from Pfizer, Daiichi Sankyo, PharmaMar, Astra Zeneca, and Roche. S.A. has acted as a consultant or advisor for Novartis and Daiichi-Sankyo. I.G. and C.P. are employees of Novartis. R.M.S. has acted as a consultant or advisor for AbbVie, Actinium, and Agios; has received personal fees from Amgen, argenx, AROG, Astellas, AstraZeneca, BioLineRx, Celgene, Cornerstone, Daiichi-Sankyo, Fujifilm, Jazz Pharmaceuticals, MacroGenics, Novartis, Ono/Theradex Oncology, Orsenix, Otsuka/Astex, Pfizer, Roche, Stemline Therapeutics, Takeda, and Trovagene; and has received institutional research support from AbbVie, Agios, AROG, and Novartis. H.D. has acted as a consultant or advisor for AbbVie, Agios, Amgen, Astellas, Astex Pharmaceuticals, Celgene, Janssen, Jazz Pharmaceuticals, Novartis, Roche, and Seattle Genetics and has received institutional research support from Amgen, AROG Pharmaceuticals, Bristol-Myers Squibb, Celgene, Jazz Pharmaceuticals, Novartis, Pfizer, and Sunesis. The remaining authors declare no competing financial interests.
Correspondence: Konstanze Döhner, Department of Internal Medicine III, University of Ulm, Albert-Einstein-Allee 23, 89081, Ulm, Germany; e-mail: konstanze.doehner@uniklinik-ulm.de.
The current affiliation for T.W.P. is Molecular Genetics Laboratory, Center for Human Genetics Laboratory, University Hospitals Cleveland Medical Center, Cleveland, OH.
The current affiliation for G.M. is Department of Hematology and Hematopoietic Cell Transplantation, Gehr Family Center for Leukemia Research, City of Hope, Duarte, CA.
The current affiliation for J.K. is Medizinische Klinik III, Städtisches Klinikum Braunschweig, Braunschweig, Germany
The current affiliation for J.M.B. is Divsion of Hematology, Department of Medicine, University of Alberta, Edmonton, AB, Canada.
The current affiliation for B.C.M. is Roche/Genentech, Basel, Switzerland.
The current affiliations for R.F.S. are Trial Center, National Center of Tumor Diseases, German Cancer Research Center Heidelberg, Heidelberg, Germany and Department of Internal Medicine V, Heidelberg University Hospital, Heidelberg, Germany.
REFERENCES
Author notes
K.D. and C.T. contributed equally to this work.
R.M.S., H.D., and C.D.B. share senior authorship.