High prevalence of IDH mutations in seronegative rheumatoid arthritis (RA) with myeloid neoplasm, elevated 2-hydroxyglutarate, dysregulated innate immunity, and proinflammatory microenvironment suggests causative association between IDH mutations and seronegative RA. Our findings merit investigation of IDH inhibitors as therapeutics for seronegative IDH-mutated RA.
TO THE EDITOR:
Autoimmune manifestations including autoimmune rheumatic disease (AIRD) are reported in 8% to 30% of patients with myeloid neoplasm (MN), including myelodysplastic syndrome (MDS), chronic myelomonocytic leukemia (CMML), acute myeloid leukemia (AML), and myeloproliferative neoplasms (MPNs).1-4 Furthermore, the risk of developing MN is elevated in autoimmune diseases.5,6 However, the mechanisms underpinning the association between autoimmune disease and MN remain unclear.
We assessed the burden of AIRD in patients with MN (n = 1702) including MDS (n = 861), AML (n = 640), MDS/MPN (n = 112), and MPN (n = 89). AIRD was identified using the International Classification of Diseases, which was then verified by experienced rheumatologists (supplemental Figure 1A-B and supplemental Table 1, available on the Blood website). The study was approved by the respective ethics committees and performed in accordance with the Declaration of Helsinki. Methodological details are provided in the supplemental materials.
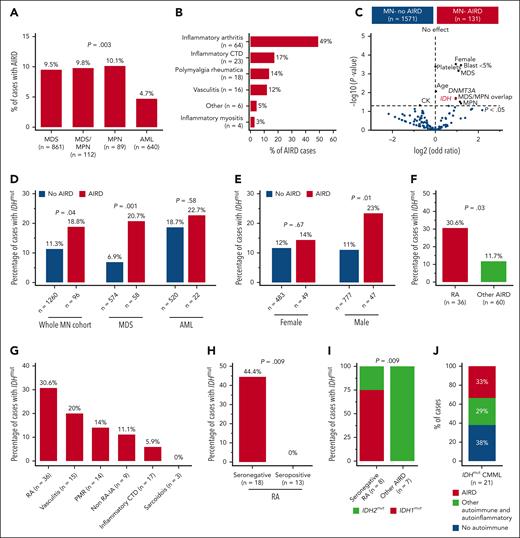
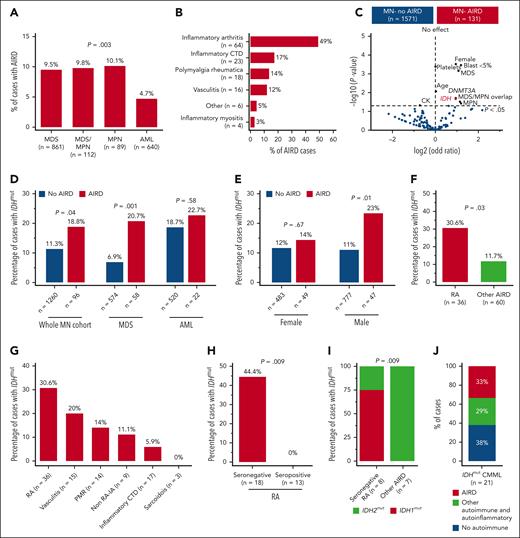
The median age of the cohort was 68 years (interquartile range, 59-75), 1023 (60%) of the subjects were male (supplemental Table 2), and 7.7% of patients with MN had AIRD (MN-AIRD). Enrichment of AIRD was observed in MDS, MDS/MPN, and MPN compared with AML (9.5%, 9.8%, 10.1%, and 4.7%, respectively; P = .003) (Figure 1A). MN cohort displayed male predominance (60.1% vs 39.9%), but a higher proportion of females had concomitant MN-AIRD (55.7% vs 44.3%; P < .01). This was confirmed when analyzed by sex (11% female vs 6% male, P < .0001), especially in MDS-AIRD (supplemental Figure 1C-E). Inflammatory arthritis (n = 64, 48.9%), inflammatory connective tissue diseases (n = 23, 17.2%), polymyalgia rheumatica (n = 18, 13.7%), and vasculitis (n = 16, 12.2%) were the most prevalent AIRD (Figure 1B). Notably, 85.9% of inflammatory arthritis diseases were rheumatoid arthritis (RA) (n = 55) followed by peripheral (n = 8, 12.5%) and axial (n = 1, 1.6%) spondyloarthropathies.
High prevalence of IDH mutation in myeloid neoplasm with RA. (A) In all, 9.5% of cases with MDS had AIRD compared with only 4.7% of cases with AML (P = .003). (B) Inflammatory arthritis is the most prevalent AIRD in the MN-AIRD cohort. Inflammatory arthritis includes RA plus peripheral spondyloarthropathy. (C) Volcano plot comparing the blood counts, bone marrow blast, cytogenetic changes, and somatic mutation profile of MN-AIRD vs MN without AIRD. (D) Enrichment of IDH mutations in MN-AIRD compared with MN without AIRD, especially MDS-AIRD compared with MDS without AIRD. (E) Unexpected enrichment of IDH mutation observed in male cases with MN-AIRD but not in female cases with MN-AIRD. (F) High burden of IDH mutation in MN with RA compared with MN with other subtype of AIRD. (G) Frequency of IDH mutation according to subtype of AIRD in MN. (H) Strikingly high frequency of IDH mutation in seronegative RA compared with seropositive RA. (I) Majority of seronegative RA harbors IDH1 mutation, in contrast to IDH2 mutation in other MN-AIRDs. (J) In an independent validation cohort, one-third of patients with IDHmut CMML have AIRD. The χ2 test was used to determine associations between categorical variables. CTD, connective tissue diseases.
High prevalence of IDH mutation in myeloid neoplasm with RA. (A) In all, 9.5% of cases with MDS had AIRD compared with only 4.7% of cases with AML (P = .003). (B) Inflammatory arthritis is the most prevalent AIRD in the MN-AIRD cohort. Inflammatory arthritis includes RA plus peripheral spondyloarthropathy. (C) Volcano plot comparing the blood counts, bone marrow blast, cytogenetic changes, and somatic mutation profile of MN-AIRD vs MN without AIRD. (D) Enrichment of IDH mutations in MN-AIRD compared with MN without AIRD, especially MDS-AIRD compared with MDS without AIRD. (E) Unexpected enrichment of IDH mutation observed in male cases with MN-AIRD but not in female cases with MN-AIRD. (F) High burden of IDH mutation in MN with RA compared with MN with other subtype of AIRD. (G) Frequency of IDH mutation according to subtype of AIRD in MN. (H) Strikingly high frequency of IDH mutation in seronegative RA compared with seropositive RA. (I) Majority of seronegative RA harbors IDH1 mutation, in contrast to IDH2 mutation in other MN-AIRDs. (J) In an independent validation cohort, one-third of patients with IDHmut CMML have AIRD. The χ2 test was used to determine associations between categorical variables. CTD, connective tissue diseases.
Enrichment for somatic mutations in the epigenetic modifiers DNMT3A (odds ratio 1.93, P = .02) and IDH1/2 (odds ratio 1.9, P = .02) was observed in MN-AIRD (Figure 1C). To investigate this observed link, we analyzed an expanded cohort of 1356 patients with MN screened for IDH1/2 mutations. Enrichment of IDH1/2 mutations was identified in all MN-AIRD cases compared with non-AIRD cases (18.8% vs 11.3%; P = .04) driven primarily by the enrichment noted in MDS-AIRD (20.7% vs 6.9%; P = .001), suggesting a link between these somatic mutations and autoimmune features (Figure 1D). A similar trend was observed in AML (22.7% vs 18.7%; P = .58) but did not reach statistical significance in part due to the higher prevalence of seropositive RA in AML compared with MDS (78% vs 29%; P = .03), and the lack of association of seropositive RA with IDH mutation. Unexpectedly, males with MN-AIRD displayed IDH1/2 mutations enrichment when compared with those without AIRD (23% vs 11%; P = .01), which was not apparent in females (14% vs 12%; P =.67) (Figure 1E).
We next evaluated AIRD subtypes in patients with IDH1/2 mutations. IDH1/2 mutations were increased in MN with RA vs other AIRD (30.6% vs 11.7%; P = .03) (Figure 1F-G) with striking enrichment in seronegative vs seropositive RA (44.4% vs 0%; P = .009) (Figure 1H). Although IDH1/2 mutations occurred with similar frequency in AIRD, we found enrichment of IDH1 mutations in seronegative RA (75% IDH1 vs 25% IDH2), and IDH2 mutations were prevalent in other AIRD (100% IDH2 vs 0% IDH1; P = .009) (Figure 1I). The higher frequency of AIRD was verified in an independent validation cohort of patients with CMML (n = 21) who were selected based on IDH mutation status, irrespective of AIRD status. Of the patients with IDH1/2-mutated CMML, 62% (n = 13) had AIRD (n = 7) or other autoinflammatory manifestations (n = 6) (Figure 1J). Our findings indicate enrichment of IDH1 and IDH2 mutations in AIRD, particularly seronegative RA.
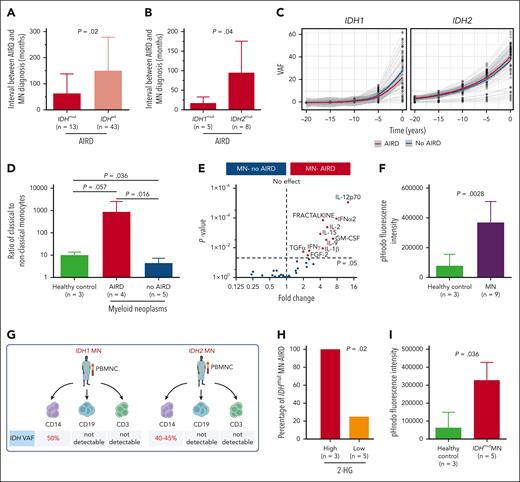
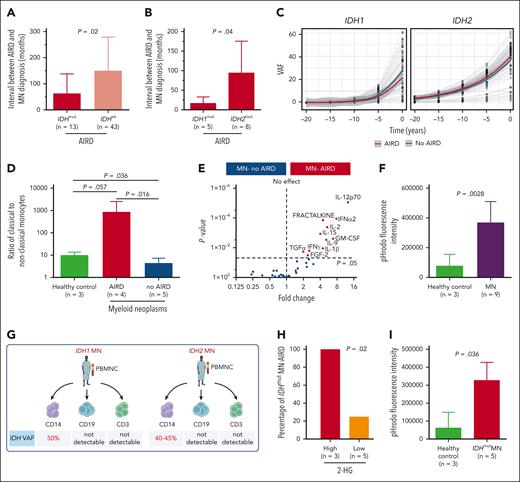
If IDH1/2 mutations play a pathogenic role in AIRD, it is plausible that mutant clones precede AIRD diagnosis. We therefore analyzed the temporal events surrounding mutation and disease onset. In 84% (n = 65) and 5% (n = 4) of cases with available information (n = 78), MN was diagnosed >12 and 1 to 12 months after the AIRD diagnosis, respectively, and in 6% (n = 5) and 5% (n = 4) of cases, MN was diagnosed within 1 month and >1 month before the AIRD diagnosis, respectively. The interval between diagnosis of RA and MN (latency) tended to be shorter in seronegative vs seropositive RA (34.6 vs 135.4 months; P = .06) (supplemental Figure 1F). Importantly, the interval from AIRD to MN was markedly shorter in IDH1/2-mutant cases compared with IDH wild-type cases (44.4 vs 106.3 months; P = .02) (Figure 2A) and even shorter in patients with IDH1 vs IDH2 (12.9 vs 85.6 months; P = .04) (Figure 2B). This short latency between AIRD and MN could be explained by the clonal expansion rate of 20% and 10% per year of IDH1- and IDH2-mutated clones, respectively, which is faster than the yearly expansion rate of 5% for DNMT3A and TP53 clones.7 Furthermore, extrapolation of the IDH1/2-mutant clonal burden prior to MN diagnosis indicates that the majority of clones are expected to be detectable at the time of AIRD diagnosis (Figure 2C).
IDH-mutant clones are likely to be present prior to diagnosis of autoimmune disease. (A) Interval between AIRD and MN diagnosis was shorter in IDH-mutated MN-AIRD compared with IDH wild-type MN-AIRD particularly in (B) IDH1-mutated compared with IDH2-mutated MN-AIRD. (C) Locally estimated scatterplot smoothing (LOESS) analysis to calculate clonal expansion kinetics of IDHmut and to evaluate if IDH clones were present at the time of AIRD diagnosis. Within each “window,” a weighted average was calculated, and the sliding window passes along the x-axis. The shaded area indicates the 95% confidence interval. Gray lines represent patients. (D) Aberrantly high ratio of proinflammatory classical to nonclassical monocytes in patients with MN-AIRD (n = 4) compared with patients with MN without AIRD (n = 5) and age-matched healthy controls (n = 3). (E) Proinflammatory cytokines secreted by innate immune cells including GM-CSF, IL-12, fractalkine, IL-15, and IL-1β were significantly higher in bone marrow plasma of MN-AIRD (n = 9) compared with MN without AIRD (n = 109). (F) Aberrantly high macrophage phagocytic activity, assessed by pHrodo Red Staphylococcus aureus Bioparticles uptake, in subjects with MN (n = 9) compared with healthy controls (n = 3). (G) Schema of IDH mutational status interrogation in various cell populations (CD14+ monocytes, CD19+ B cells, and CD3+ T cells) in 2 patients with IDH mutation. (H) All IDHmut MN with high bone marrow plasma level of 2-HG had AIRD compared with only 25% of cases with low 2-HG level. (I) Aberrantly high in vitro macrophage activity in subject with IDHmut MN (n = 5) compared with healthy control (n = 3). All bars indicate mean, and all error bars indicate SD. The Mann-Whitney test was used to detect statistically significant differences between cohorts. The χ2 test was used to determine associations between categorical variables. FGF-2, fibroblast growth factor 2; TGFα, transforming growth factor α; GM-CSF, granulocyte-macrophage colony-stimulating factor; IFN-α2, interferon α2; IFNγ, interferon γ; VAF, variant allele frequency.
IDH-mutant clones are likely to be present prior to diagnosis of autoimmune disease. (A) Interval between AIRD and MN diagnosis was shorter in IDH-mutated MN-AIRD compared with IDH wild-type MN-AIRD particularly in (B) IDH1-mutated compared with IDH2-mutated MN-AIRD. (C) Locally estimated scatterplot smoothing (LOESS) analysis to calculate clonal expansion kinetics of IDHmut and to evaluate if IDH clones were present at the time of AIRD diagnosis. Within each “window,” a weighted average was calculated, and the sliding window passes along the x-axis. The shaded area indicates the 95% confidence interval. Gray lines represent patients. (D) Aberrantly high ratio of proinflammatory classical to nonclassical monocytes in patients with MN-AIRD (n = 4) compared with patients with MN without AIRD (n = 5) and age-matched healthy controls (n = 3). (E) Proinflammatory cytokines secreted by innate immune cells including GM-CSF, IL-12, fractalkine, IL-15, and IL-1β were significantly higher in bone marrow plasma of MN-AIRD (n = 9) compared with MN without AIRD (n = 109). (F) Aberrantly high macrophage phagocytic activity, assessed by pHrodo Red Staphylococcus aureus Bioparticles uptake, in subjects with MN (n = 9) compared with healthy controls (n = 3). (G) Schema of IDH mutational status interrogation in various cell populations (CD14+ monocytes, CD19+ B cells, and CD3+ T cells) in 2 patients with IDH mutation. (H) All IDHmut MN with high bone marrow plasma level of 2-HG had AIRD compared with only 25% of cases with low 2-HG level. (I) Aberrantly high in vitro macrophage activity in subject with IDHmut MN (n = 5) compared with healthy control (n = 3). All bars indicate mean, and all error bars indicate SD. The Mann-Whitney test was used to detect statistically significant differences between cohorts. The χ2 test was used to determine associations between categorical variables. FGF-2, fibroblast growth factor 2; TGFα, transforming growth factor α; GM-CSF, granulocyte-macrophage colony-stimulating factor; IFN-α2, interferon α2; IFNγ, interferon γ; VAF, variant allele frequency.
AIRD are thought to be driven through adaptive immunity. However, autoinflammatory disorders such as VEXAS (vacuoles, E1 enzyme, X linked, autoinflammatory, somatic) syndrome are associated with innate immune activation and MDS.8 We therefore analyzed T-cell subsets and monocyte and macrophage function in AIRD vs non-AIRD MN. We found a higher ratio of proinflammatory CD14+CD16− classical monocytes to nonclassical anti-inflammatory CD14−CD16+ monocytes in MN-AIRD compared with age-matched controls and cases of MN without AIRD (Figure 2D; supplemental Figure 2). In contrast, no difference in T-cell subsets (including naive, central memory, effector memory, and terminally differentiated CD4+ and CD8+ T cells) was observed (supplemental Figure 3). Furthermore, we observed increased levels of proinflammatory cytokines, including granulocyte-macrophage colony-stimulating factor, interleukin-12 (IL-12), fractalkine, IL-15, and IL-1β, in the bone marrow plasma of patients with MN-AIRD vs those without AIRD (Figure 2E). Consistent with these cytokines being predominantly derived from innate immune monocyte/macrophages rather than T cells, we observed increased monocyte-derived phagocytic activity (assessed by flow cytometry) in MN (Figure 2F).
We then isolated CD14+ monocytes, CD3+ T cells, and CD19+ B cells from 2 patients with AIRD with known IDH1/2 mutations (Figure 2G). Interestingly, high VAF clones were detected in monocytes (40%-50%) but not in T cells (<1%) or B cells (<1%) from both patients (Figure 2G; supplemental Figure 4), consistent with intracellular (R)-2-hydroxyglutarate (2-HG) production in monocyte/macrophage cells rather than T cells, leading us to assess 2-HG levels in patients with AIRD. Strikingly, all 3 cases with high 2-HG (bone marrow metabolite peak abundance above median of >9.3 × 105) had AIRD compared with only 1 of the 5 cases with low 2-HG (<9.3 × 105 median abundance, P = .02) (Figure 2H). Finally, we noted significantly higher macrophage phagocytic activity in subjects with IDH1/2-mutant MN compared with controls (Figure 2I).
IDH mutations are neomorphic and lead to the production of an alternative metabolite in the citric acid cycle, 2-HG, which inhibits TET2 deoxygenase with resultant genomewide epigenetic modifications. This study demonstrates the enrichment of IDHmut in MN-AIRD, in line with previous findings.4 Importantly, our study demonstrates a strong association between IDH-mutated MN and a specific type of AIRD-seronegative RA, as well as a temporal relationship between IDH-mutant clones and AIRD. The observed elevated 2-HG in seronegative RA and IDH1/2 mutations in myeloid rather than T cells indicates a causative association between IDH1/2 mutation and AIRD. Emerging literature from in vitro9 and in vivo studies reports proinflammatory effects of IDH mutations on monocytes9 and macrophages,10 in addition to an elevated IL-18 level in individuals with IDH1 clonal hematopoiesis.11 Collectively our findings suggest a causative relationship between IDH1/2 mutations, MN, and seronegative AIRD, meriting further exploration of mutant IDH inhibitors such as ivosidenib,12 enasidenib,13 or complex I inhibitor14 as therapeutic options for seronegative RA.
Acknowledgments
The authors are grateful to the patients, their families, the Mayo Clinic Acute Leukemia and Myeloid Neoplasms Biobank, and the South Australia Cancer Research Biobank. The authors acknowledge editing support by Carla Toop.
Some of the data in this publication were produced in the Mayo Clinic Cytogenetics Core Laboratory, which is supported in part by the Mayo Clinic Comprehensive Cancer Center Grant, funded by the National Institutes of Health, National Cancer Institute (P30CA15083). L.E.H. received University of Adelaide Research Training Program Scholarship and Royal Adelaide Hospital (RAH) Research Committee Dawes Top-Up Scholarship. M.V.S. was supported by Mayo Clinic Research Pipeline K2R Transition Award and Bridget Kiely Clinician Career Development in Transplant Research and Mayo Clinic, Rochester. D.K.H. was supported by a National Health and Medical Research Council/Medical Research Future Fund Investigator Grant (MRF1195517), Cancer Australia (#2013617), and Leukemia Foundation Australia (#22007).
Authorship
Contribution: L.E.H., M.D.W., and S.P. collected and annotated autoimmune rheumatic disorders, provided rheumatology expertise, and edited the manuscript; M.K. performed cytokines/chemokines profiling, helped with analysis, and edited the manuscript; A.S. performed monocytes, macrophages, and T-cells subsets analyses and edited the manuscript; K.L. and S.S. performed and analyzed 2-HG levels assay and edited the manuscript; C.T.-P. performed IDH sequencing in isolated T cells and edited the manuscript; R.C. and J.J.L. collated clinical data of myeloid neoplasms and autoimmune rheumatic diseases and edited the manuscript; H.S.S., A.B., S.B., R.J.D., S.E.S., and C.N.H. profiled somatic mutation of the samples and edited the manuscript; D.T.Y. edited the manuscript; N.R. provided expertise in macrophage function and edited the manuscript; R.T. provided expertise on autoimmune rheumatic disease and edited the manuscript; M.P. contributed validation cohort and edited the manuscript; D.T. edited the manuscript, designed experiments, and supervised mass spectrometry analyses and IDH sequencing in isolated T cells; C.H.K. performed statistical analysis and edited the manuscript; M.V.S. contributed the patient data and edited the manuscript; and D.K.H. designed the project, secure funding, supervised, analyzed results, and wrote the manuscript.
Conflict-of-interest disclosure: M.D.W. received a research grant from GlaxoSmithKline for an unrelated research project; M.P. declares membership on an entity’s board of directors or advisory committees (Stemline Therapeutics) and research funding (Kura Oncology); M.V.S. received research funding to the institution from AbbVie, Celgene, MRKR Therapeutics, KURA Oncology, and Astellas; D.K.H. declares membership on an entity’s board of directors or advisory committees (AbbVie, Novartis). The remaining authors declare no competing financial interests.
Correspondence: Devendra K. Hiwase, Haematology Department, The Royal Adelaide Hospital, 1 Port Rd, Adelaide, SA 5000, Australia; email: devendra.hiwase@sa.gov.au.
References
Author notes
L.E.H. and M.D.W. contributed equally to this work.
C.H.K., M.V.S., and D.T. contributed equally to this work.
Additional methods and data can be found in the supplementary Data. For original data, please contact corresponding author, Devendra K. Hiwase (devendra.hiwase@sa.gov.au).
The online version of this article contains a data supplement.