Abstract
To assess the clinical significance of minimal residual disease (MRD) detection by polymerase chain reaction (PCR) we analyzed samples from 26 patients with mantle cell lymphoma (MCL) who had undergone bone marrow transplantation (BMT) at the Dana-Farber Cancer Institute. The BCL-1/IgH translocation and clonally rearranged Ig heavy chain genes (IgH) provided molecular markers for detection and follow-up of MRD by polymerase chain reaction (PCR) amplification in 19 of the 26 (73%) MCL patients studied. IgH gene sequencing analysis showed somatic mutations in MCL that are characteristic of an antigen driven process suggesting that, in MCL, the final malignant transformation occurs in a mature B cell. Of the 19 patients with a PCR amplifiable marker, 17 underwent autologous, 1 an allogeneic, and 1 a syngeneic bone marrow transplantation (BMT). All patients had PCR-detectable MRD in the bone marrow (BM) at the time of BMT, irrespective of any history of histological BM involvement. In contrast to other B-cell malignancies, we found that immunological purging with complement-mediated lysis eradicated PCR-detectable MCL in only two patients. Moreover reinfusion of MRD was associated with a poor outcome. More than half of the patients undergoing autologous BMT had relapsed by the time of restaging at 2 years after autologous BMT. In four MCL patients in whom no residual lymphoma was reinfused, including the allogeneic and the syngeneic BMT, only one patient relapsed. Persistence of MRD detection after BMT was also associated with a high probability of relapse, although one patient did not have PCR-detectable MRD in peripheral blood or BM before relapse at nodal sites. We conclude that PCR amplification of disease-specific markers is a feasible and sensitive method to assess MRD and its clinical significance in patients with MCL. Moreover, PCR amplification provides a tool to evaluate modifications of purging and stem cell collection procedures that may be required for the management of this otherwise incurable disease.
MANTLE CELL LYMPHOMA (MCL) is now clearly recognized as a distinct subtype of non-Hodgkin's lymphomas (NHL), based on its morphological, immunophenotypical, and molecular markers.1,2 MCL is characterized by advanced stage disease at presentation with frequent bone marrow (BM) infiltration.1,3-5 Although previously categorized as an indolent lymphoma, MCL has low chemosensitivity and poor disease-free survival (DFS) and appears incurable with conventional therapy.5-10 Investigators have studied the use of high-dose therapy with stem cell rescue for the management of this disease, but no large studies have determined the role of this treatment in MCL.4,7,8 10 Furthermore, this approach has been hampered by the frequent BM involvement in MCL, limiting the application of autologous bone marrow transplantation (ABMT). At the Dana-Farber Cancer Institute (Boston, MA) immunological purging of autologous BM is used in an attempt to eradicate contaminating tumor cells. For this reason, patients with MCL with BM involvement have been eligible for high-dose therapy and ABMT. Purging is performed by using a cocktail of anti–B-cell monoclonal antibodies (MoAbs) and complement-mediated lysis.
MCL is characterized molecularly by the translocation t(11; 14), which fuses the BCL-1 gene on chromosome 11 with the immunoglobulin heavy chain (IgH) locus on chromosome 14.11,12 This translocation alters the expression of the putative oncogene cyclin D1. This is not by itself tumorigenic and further transformational processes appear to be necessary for the development of MCL.1,13 The BCL-1/IgH translocation occurs in 70% to 100% of MCL patients when analyzed by multiple probing or fluorescent in situ hybridization.14,15 Up to 50% of such translocations cluster at a region known as the major translocation cluster (MTC) region, making this an ideal target for polymerase chain reaction (PCR) amplification and minimal residual disease (MRD) detection.15-18 In addition, as occurs with other B-cell malignancies, MCL expresses clonally rearranged IgH genes. This provides an additional disease marker that can be PCR amplified to detect MRD.19-21 We have previously assessed the clinical significance of PCR detection of MRD at the time of and after ABMT in other B-cell malignancies, including follicular lymphoma and B-cell chronic lymphocytic leukemia (CLL).20-24 In these studies, we showed that immunological purging of autologous BM was capable of eradicating PCR-detectable disease in up to 50% of the patients. Eradication of PCR-detectable MRD from the purged BM23 and the patient samples after ABMT20,21,24 were associated with improved disease-free survival (DFS). Although several studies have examined PCR detection of t(11; 14) in MCL,15-18 only a single case report published to date has examined the role of PCR amplification to assess MRD after therapy.25 PCR amplification and direct sequencing of the IgH gene also permits study of IgH gene variable region (VH) usage and assessment of somatic mutations from the germline configuration. In other B-cell malignancies, including multiple myeloma and follicular lymphoma, nonrandom VH gene usage and nonrandom replacement mutations that occur in excess in the complementarity-determining region (CDR) sequences have led to the conclusion that the final tumorigenic event happens in a mature B cell after antigen selection.26-29 In contrast, previous studies, albeit in small numbers of patients, have not identified somatic mutations in MCL, suggesting that these lymphoma cells are not antigen driven.30 31
In the present study we examined MRD by PCR amplifying BCL-1/IgH translocations and IgH rearrangements in 26 patients with MCL who have undergone BMT at our institution. We obtained molecular markers of disease for MRD detection in 19 of these 26 patients (73%). In contrast to previously published studies,30 31 we observed a significant clustering of replacement mutations in the CDRs of the VH genes in MCL. This suggests that the final tumorigenic event may have occurred in a mature B cell and that the process may be, in part, antigen driven. We used PCR assessment of MRD to evaluate the efficacy of immunological purging. Unlike our findings in other B-cell malignancies, including follicular lymphoma and B-cell CLL, we observed that immunological purging failed to eradicate PCR-detectable disease in the vast majority of patients with MCL. Moreover, reinfusion of PCR detectable MRD in MCL appeared to be associated with an unfavorable outcome after ABMT. Persistence of PCR-detectable MRD in BM and/or peripheral blood (PB) was associated with a high probability of relapse. We concluded that PCR amplification is a feasible and sensitive method to assess the clinical significance of MRD in patients with MCL undergoing BMT and that immunological purging as performed in our previous studies is insufficient to eradicate this disease.
MATERIALS AND METHODS
Patients. A group of 26 lymphoma patients diagnosed with MCL who have undergone BMT at the Dana-Farber Cancer Institute were studied. All patients were classified as having MCL after central review of histology at the Histopathology Department at Brigham and Women's Hospital. Patients were considered candidates for BMT based on the presence of progressive disease at the time of initial presentation or at relapse. All patients had chemosensitive disease as defined by their ability to achieve a protocol eligible disease state with the largest nodal mass no greater than 2 cm in largest diameter and BM infiltration less than 10% of the intertrabecular space before proceeding to BMT. The harvested marrow of patients undergoing autologous BMT was purged by using anti–B-cell MoAbs and complement-mediated lysis. Patients' autologous BM was treated by using B1 (anti-CD20) in initial studies and later with a cocktail of anti–B-cell containing B1, J5 (anti-CD10). To attempt to increase the efficacy of purging, B4 (anti-CD19) was added to the cocktail of MoAbs used for purging. Human leukocyte antigen (HLA) identical sibling donor marrow for the patient who underwent allogeneic BMT was T-cell depleted by using T12 (anti-CD6) MoAb and complement-mediated lysis. The syngeneic marrow was infused after harvest without processing. These investigative protocols were approved by the Institutional Review Board of Dana-Farber Cancer Institute (Boston, MA) and all patients gave informed consent.
DNA preparation. BM and PB samples were obtained before, at the time of, and after BMT. The harvested BM samples were analyzed before and after immunological purging. PB and BM mononuclear cells were isolated with Ficoll gradient centrifugation (Pharmacia, Uppsala, Sweden). Thereafter, cells were lysed and DNA extracted as previously described.32 Before PCR amplification DNA samples were heated to 96°C (10 minutes) to denature proteinase K.
PCR amplification of BCL-1/IgH translocation. PCR amplifications were performed on a GeneAmp PCR System 9600 or DNA Thermal Cycler 480 (Perkin Elmer, Branchburg, NJ). DNA from diagnostic samples was amplified by semi-nested PCR. The first amplification used a sense 5′ primer, 5′ CTGAAGGACTTGTGGGTTGC 3′ located upstream of the major translocation cluster (MTC) on the BCL-1 gene (240 bp upstream of the Sac I site).15,16 The second PCR reaction used a sense 5′ primer, 5′ TAAGGCTGCTGTACACATCG 3′, located 43 bp downstream from the first primer. Both amplifications were performed with an antisense 3′ joining region (JH) consensus primer 5′ ACCTGAGGAGACGGTGACC 3′. PCR was performed in a final volume of 50 μL by using PCR buffer (50 mmol/L KCl, 10 mmol/L TrisCl, 2.25 mmol/L MgCl2 , 0.01% gelatin), 50 μmol/L of each dNTP (Ultrapure dNTP Set; Pharmacia, Lund, Sweden), 200 nmol/L of each primer, 1.5 U Amplitaq DNA polymerase (Perkin Elmer), ddH2O, and 1 μg target DNA for the first amplification or 2 μL of the initial PCR product for the second amplification. Reaction conditions for the first and second round amplification were identical: 30 cycles with denaturation at 94°C (first cycle for 3 minutes, thereafter for 30 seconds), annealing at 55°C for 30 seconds and extension at 72°C for 30 seconds, with a final extension time of 10 minutes. Twenty microliters of the final PCR product was electrophoresed in 3% agarose gel containing ethidium bromide and visualized under ultraviolet (UV) light. Each reaction was performed by using appropriate negative and positive controls. BCL-1/IgH sequence analysis was performed by the Molecular Core Facility at the Dana-Farber Cancer Institute, with Perkin Elmer Applied Biosystems AB 373A DNA Sequencer (Perkin Elmer). Patient sequences were aligned to the BCL-1 gene15 and breakpoint sites located by using GeneJockey II (Biosoft, Cambridge, UK). The N inserts and JH regions of the BCL-1/IgH translocation were identified by BLAST Search network service (National Center for Biotechnology Information, Bethesda, MD). Specificity was determined by using DNA from normal donor PB as negative control and DNA from a cell line that has the BCL-1/IgH translocation as positive control. Positive control DNA was a kind gift from Dr R. Rimokh, Hopital Edouard Herriot, Lyon, France. The sensitivity of detection of MRD was assessed by PCR amplification of 10-fold serial dilutions of tumor cell DNA into normal donor PBL DNA. PCR amplification of the major breakpoint region (MBR) and the minor cluster region (mcr) of the BCL-2 translocation was performed as previously described.32
Allele-specific oligonucleotide amplification of clonal IgH rearrangement. To identify clonal IgH rearrangements we PCR-amplified diagnostic samples using either seven family specific framework region one (FR1) VH primers19 and JH consensus primer or an FR3 VH consensus primer (VHcon) and the JH consensus primer, as previously described.20 PCR reactions were performed in 100 μL final volume by using PCR buffer II, 2.5 mmol/L MgCl2, 200 nmol/L each primer, 200 μmol/L of each of dATP, dTTP, dGTP, dCTP, 2.5 U AmpliTaq DNA polymerase (Perkin Elmer), 1 μg target DNA, and ddH2O. Samples were amplified “hot start” with AmpliWax GEM 100 beads (Perkin Elmer) for 30 cycles with denaturation at 94°C (first cycle for 3 minutes, thereafter for 30 seconds), annealing at 62°C for 30 seconds, and extension at 72°C for 30 seconds with a final extension time of 10 minutes. Twenty microliters was electrophoresed in 3% agarose gel containing ethidium bromide and visualized under UV light. Clonal amplification gave a sharp band of the appropriate size of approximately 100 bp with the VHcon and 350 bp for the VH family primers.
The PCR amplified clonal product was electrophoresed, excised, and purified using Wizard PCR preps (Promega, Madison, WI). The DNA content was measured by optical density (OD) and approximately 700 ng used for sequencing. The relevant VH family primer and JH consensus primer were used for the direct sequencing performed as above. Sense and antisense sequences were aligned and made contiguous with GeneJockey II. VH-, D-, and JH-regions and N-inserts were identified by BLAST. The CDR III region was identified as the junction of these three regions including the N-inserts. 20mer antisense allele-specific oligonucleotides (ASO) primers were designed from the CDR III regions including N-insert. All oligonucleotides were designed to have a melting temperature of approximately 60°C. Oligonucleotides were synthesized by a PE AB 3948 DNA Synthesizer (Perkin Elmer) in the Molecular Core Facilities, Dana-Farber Cancer Institute.
DNA from the patient samples was amplified by semi-nested PCR. The first amplification used the relevant VH family and JH consensus primers. A 2-μL aliquot of the initial PCR product was used as the template for the second amplification using the relevant VH family primer (sense) and the designed ASO primer (antisense). PCR reaction conditions were essentially the same as described previously for the BCL-1/IgH translocation. The annealing temperature for the first amplification was 62°C and 5°C below the melting temperature (TM-5°C) for the designed ASO primer in the second round. This annealing temperature was frequently adjusted until background was abolished and sensitivity still acceptable. Specificity was determined by using the diagnostic sample as positive control and BM samples from three other MCL patients as negative controls. The sensitivity of detection of MRD was assessed by PCR amplification of 10-fold serial dilutions tumor cells into normal donor PBL.
Assignment of somatic mutation in the VH region. VH region sequences were assessed as described previously and were derived from at least two 100% aligned sequences. Mutations were identified by comparing every VH region obtained to the best aligned VH germline gene with BLAST. FR1-3 and CDR I to II regions were identified with the aid of Kabat et al.33 The appropriate reading frame was identified with Strider 1.2 (Marck, Service de Biochimie et de Genetique Moleculaire, Gif-sur-Yvette, France) to determine if an exchange mutation was silent, with no alteration in the deduced amino acid sequence, or a replacement mutation that changed the deduced amino acid sequence.
Statistical analysis. Binomial probability (Pr) analysis was performed according to a previously published model34,35 to determine whether replacement mutations at the CDRs were likely to have occurred by chance or whether there was significant excess of replacement mutations in the CDRs, suggesting that antigen-driven selection had occurred. We used the analysis Pr (×replacement mutations in the CDRs) ∼Bi(n,p), and calculated exact tail probabilities. N is the total number of mutations. The number of replacement mutations occurring in the FRs was multiplied by two to account for the fact that some FR replacement mutations might result in deleterious mutations and result in loss of the clone.34 P is the product of the relative length of the CDRs and the relative number of replacement mutation occurring in the CDRs. Fisher's exact test was used to evaluate VH use in MCL and compare purging efficacy in MCL and B-cell CLL.
RESULTS
PCR amplifiable molecular marker of disease in MCL. BM and PB samples from 26 patients with MCL who have undergone BMT at the Dana-Farber Cancer Institute were studied. All patients had chemosensitive disease at the time of BMT and had achieved a protocol eligible disease state as described previously. Lymph node biopsy material from all cases was reviewed centrally and confirmed the diagnosis of MCL.
We first assessed whether a molecular marker of disease could be PCR amplified from diagnostic tissue from these 26 MCL patients. Although the majority of patients with MCL would be expected to have a BCL-1/IgH, not all cases can be PCR amplified because of considerable heterogeneity in the site of translocation at the BCL-1 locus. Semi-nested PCR amplification was performed with primers spanning the MTC locus of the BCL-1/IgH translocation. With this approach a PCR product was identified in 9 of these 27 patients (33%) varying in size from 300 bp to 550 bp. Sequence analysis showed that in all cases the PCR product contained sequences from the BCL-1 region and the JH region of the IgH with an insert juxtaposed between. The size of this N insert was in most cases approximately 10 bp; however, in patient no. 1826 there was a large insert of 250 bp between the identified BCL-1 and JH sequences (data not shown). This insert did not align to known sequences in the database and, therefore, was considered an N-insert, although this possibly represents a case of a complex translocation with the unidentified sequence arising from another unknown chromosomal site. Specificity of PCR amplification of the BCL-1/IgH translocation was optimized by using appropriate negative and positive controls. The limit of detection of MRD for PCR amplification of BCL-1/IgH translocation was reproducible at the level of 10−5 (data not shown).
We also attempted to PCR amplify and sequence the IgH rearrangements from BM samples obtained at the time of referral for BMT from these 26 patients. A clonal band was PCR amplified and sequenced in 16 of the 26 patients (61%) by using one of the seven family specific VH FR1 primers and the JH consensus primer. In contrast to our previous experience in CLL,20 a clonal product could be PCR amplified and sequenced by using the degenerate VH consensus primer in only one patient's diagnostic sample (no. 1826). The clonal PCR product of the malignant clone, acquired with the relevant VH family specific primer, was gel purified and directly sequenced. From the clonal IgH sequence we identified the VH, D, and JH regions and the N-inserts of the CDR III. ASOs were designed from the CDR III. Specificity and sensitivity were tested as described in the Materials and Methods. The limit of detection of MRD using the ASO primers ranged from 10−3 to 10−5 dependent on the specificity of the ASO primer that could be obtained in each patient. In general, the larger the N insert, the more specific the primer and the higher the sensitivity of detection. VH family and JH usage of IgH rearrangements of the MCL patients are shown in Fig 1. As can be seen, VH4 was relatively over-represented and was used in 47% of cases, as commonly as the much larger VH3 family (47%). VH1 was used in only one patient (6%) whereas VH2, VH5, and VH6 were not represented. In Fig 2 the CDR III sequences from these patients are shown with the sequences used to construct suitable ASO primers outlined.
Rearranged Ig heavy chain gene variable region (VH) family and joining region (JH) usage in MCL.
Rearranged Ig heavy chain gene variable region (VH) family and joining region (JH) usage in MCL.
Complementarity determining region III (CDR III) from MCL patients undergoing BMT. VH, D, and JH regions and N-inserts from the CDR III are shown. Location of allele specific oligonucleotide (ASO) primers are shown in bold. VH, D region (where it could be identified) and JH usage for each patient is noted.
Complementarity determining region III (CDR III) from MCL patients undergoing BMT. VH, D, and JH regions and N-inserts from the CDR III are shown. Location of allele specific oligonucleotide (ASO) primers are shown in bold. VH, D region (where it could be identified) and JH usage for each patient is noted.
Nine patients had a PCR amplifiable BCL-1/IgH translocation, and in 16 patients a clonal IgH product could be PCR amplified and sequenced. Six patients had both a PCR amplifiable BCL-1/IgH translocation and clonal IgH rearrangement. Therefore, by using this combined strategy a clonal product could be PCR amplified in 19 of the 26 patients (73%) with MCL. The clinical characteristics of these 19 patients are shown in Table 1. Seventeen of these patients underwent ABMT. One patient underwent T-cell depleted allogeneic BMT from an HLA-identical sibling donor and one a syngeneic BMT. Three patients (no. 1676, 1846, and 1863) underwent BMT as consolidation therapy after initial induction therapy, although only one of these patients (no. 1846) achieved complete remission (CR) to front line therapy. All other patients underwent BMT after subsequent disease progression. All of these patients received salvage chemotherapy and all had chemosensitive disease at the time of BMT. Of note only one of these patients (no. 1917) had ever achieved CR and all other patients never achieved a CR to either induction or salvage therapy before preceding to BMT.
Somatic mutations in MCL IgH rearrangements. The entire VH region encompassing FR 1 to 3 and CDR I to II regions were sequenced on both strands from samples obtained at multiple time periods from 10 patients. The deduced amino acid sequences from these VH regions are shown in Fig 3. FRs and CDRs were identified and compared with the germline configuration to identify somatic mutations. As can be seen in Fig 3, 5 patients (50%) exhibited germline configuration and had no somatic mutations in the VH regions. Somatic mutations in the VH regions were identified in the other 50%. In these, four silent and three replacement mutations were found within the FRs, whereas one silent and seven replacement mutations were found within the CDRs. Binomial probability analysis, including all 10 patients, was performed as described previously. This analysis showed that the occurrence of replacement mutations in the CDRs was significantly greater than expected if the event was occurring randomly (P < .05). This suggests that these mutations may have arisen by an antigen-driven selection in MCL.
Deduced amino acid sequences of the VH regions of tumor cells from patients with mantle cell lymphoma. Comparisons are made with the best aligned germline VH genes. The first five patients had VH regions identical to (=) germline VH genes. In the latter five cases, the best aligned germline VH genes are fully shown. Dots indicate alignment between the patient VH region and the germline VH gene. Exchange mutations are indicated by the letter of the derived amino acid. Replacement mutations shown in uppercase and silent mutations in lowercase. The CDR regions are underlined. Germline (GL) VH gene sequences derived from the data-bases as shown: GL1 = EMB‖X92278‖HSIGVH5. GL2 = EMB‖Z14237‖HSVH433G. GL3 + 4 = GB‖L10090‖HUMIGHCAC. GL5 = GB‖M99406‖HUM10IGVH. GL6 = EMB‖X92278‖HSIGVH5. GL7 = GB‖UO3893‖HSU03893. GL8 = GB‖L06618‖HUMIGHHVAAE. GL9 = GB‖L10094‖HUMIGHCAG. GL10 = GB‖L10088‖HUMIGHCAA.
Deduced amino acid sequences of the VH regions of tumor cells from patients with mantle cell lymphoma. Comparisons are made with the best aligned germline VH genes. The first five patients had VH regions identical to (=) germline VH genes. In the latter five cases, the best aligned germline VH genes are fully shown. Dots indicate alignment between the patient VH region and the germline VH gene. Exchange mutations are indicated by the letter of the derived amino acid. Replacement mutations shown in uppercase and silent mutations in lowercase. The CDR regions are underlined. Germline (GL) VH gene sequences derived from the data-bases as shown: GL1 = EMB‖X92278‖HSIGVH5. GL2 = EMB‖Z14237‖HSVH433G. GL3 + 4 = GB‖L10090‖HUMIGHCAC. GL5 = GB‖M99406‖HUM10IGVH. GL6 = EMB‖X92278‖HSIGVH5. GL7 = GB‖UO3893‖HSU03893. GL8 = GB‖L06618‖HUMIGHHVAAE. GL9 = GB‖L10094‖HUMIGHCAG. GL10 = GB‖L10088‖HUMIGHCAA.
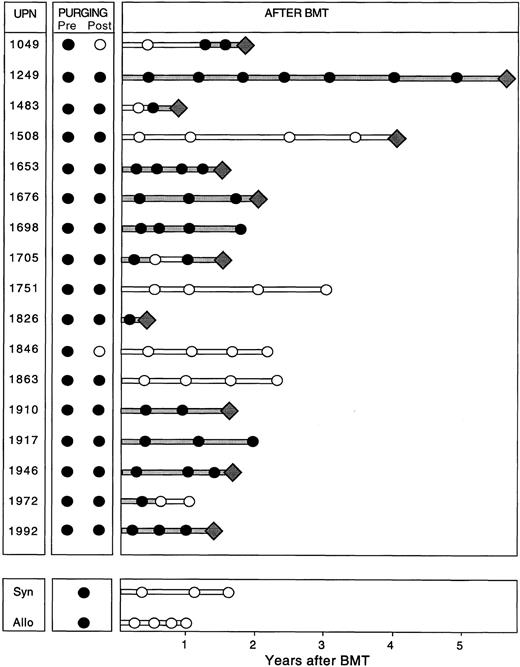
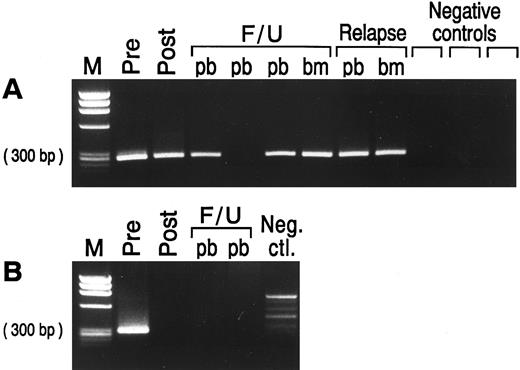
PCR assessment of immunologic purging. Of the 19 patients with PCR amplifiable markers at hand, 5 had no morphological evidence of disease in the BM at the time of BMT (Table 1). However, all 19 patients had PCR-detectable disease in the BM at this time, irrespective of whether the BM was involved by morphological assessment. The BM from patients undergoing autologous BMT were harvested and purged as described previously. After three cycles of immunological purging, no detectable B cells were detected by fluorescence-activated cell sorting (FACS) scan analysis in any of the patients (data not shown). Samples obtained before and after immunological purging were available for analysis in all 17 patients undergoing ABMT. Representative results of PCR analysis performed before and after immunological purging are shown in Fig 4A, illustrating an example in which PCR-detectable lymphoma cells remain after immunological purging, and in Fig 4B, showing successful immunological purging and eradication of PCR detectable MRD. The results of PCR amplification of all patient samples before and after purging are shown in Fig 5. As can be seen, purging successfully eradicated PCR-detectable MRD in only two (no. 1049 and 1846) patients undergoing ABMT. Of note, neither of these patients had any history of morphological infiltration of the BM, although both had PCR evidence of disease in their harvested BM. In the remaining 15 patients (88%) residual lymphoma cells were still detectable after purging. In all 5 patients in whom both BCL-1/IgH translocation and IgH rearrangement markers were available, identical results before and after purging were obtained using both methods. Of the 4 patients who received BM with no PCR-detectable MRD (no. 1049 and 1846 after ABMT, and the allogeneic and syngeneic BMT) only 1 has relapsed whereas the other 3 have no PCR evidence of MRD after BMT. Of the 15 patients reinfused with BM containing PCR-detectable lymphoma cells, 10 patients have relapsed and 2 patients have persistence of PCR-detectable MRD after ABMT.
EtBr stained agarose gel with semi-nested PCR products from prelysis and postlysis BM harvest and from peripheral blood (pb) and/or bone marrow (bm) samples at time of BMT, post-BMT follow-up (F/U), and relapse in patients with MCL. (A) Patient no. 1705 samples tested with the relevant ASO primer. The ASO primer specific for patient no. 1705 did not amplify three different MCL patients used as negative controls. (B) Patient no. 1846 samples tested with the BCL-1/IgH primers. No clonal product was seen in the normal donor PBL used as negative control. Negative control, neg. ctl.; M, molecular weight marker.
EtBr stained agarose gel with semi-nested PCR products from prelysis and postlysis BM harvest and from peripheral blood (pb) and/or bone marrow (bm) samples at time of BMT, post-BMT follow-up (F/U), and relapse in patients with MCL. (A) Patient no. 1705 samples tested with the relevant ASO primer. The ASO primer specific for patient no. 1705 did not amplify three different MCL patients used as negative controls. (B) Patient no. 1846 samples tested with the BCL-1/IgH primers. No clonal product was seen in the normal donor PBL used as negative control. Negative control, neg. ctl.; M, molecular weight marker.
Results of semi-nested PCR analysis on samples obtained at harvest (Pre), after purging (Post), and after ABMT and at the time of and after syngeneic (Syn) or allogeneic (Allo) BMT in patients with MCL. Filled lines indicate continuous PCR-positive results and unfilled lines indicate continuous PCR-negative results. The diamonds represent relapse.
Results of semi-nested PCR analysis on samples obtained at harvest (Pre), after purging (Post), and after ABMT and at the time of and after syngeneic (Syn) or allogeneic (Allo) BMT in patients with MCL. Filled lines indicate continuous PCR-positive results and unfilled lines indicate continuous PCR-negative results. The diamonds represent relapse.
The overall poor results obtained after immunological purging with MCL are in marked contrast to those seen after purging patients with follicular lymphoma23 or CLL.20 With these diseases 50% of autologous marrow samples could be successfully purged of PCR-detectable disease by the same protocol. When we compared the efficacy of purging in these 17 patients with MCL to that of 46 patients with CLL who have undergone ABMT, there was a statistically significant decrease in the efficacy of purging in MCL compared with CLL (P = .007). Both diseases are malignancies of CD19+/CD5+ cells, and the efficacy of purging was largely assessed by PCR analysis of IgH rearrangements.
Detection of minimal residual lymphoma cells after BMT. Representative results of PCR amplification of follow-up samples after BMT are shown in Fig 4, with PCR products obtained employing the ASO primer (Fig 4A) and the BCL-1 primers (Fig 4B), respectively. The results of PCR analysis of all samples obtained after BMT are shown in Fig 5. As can be seen, 11 of the 17 patients undergoing ABMT (65%) relapsed after ABMT. Sequence analysis showed that the relapse clone was identical to that obtained at the time of initial presentation. Nine patients had relapsed by the time of their 2-year follow-up visit after ABMT, whereas two additional patients relapsed at 4 years (no. 1508) and almost 6 years (no. 1249) after ABMT. Nine patients had persistence of PCR-detectable MRD in all samples obtained after ABMT and seven of these patients (78%) relapsed. In patients no. 1049, 1483, and 1705 no PCR-detectable disease could be found in BM or PB samples obtained at early time points after ABMT, but all other samples contained PCR-detectable disease and all three patients subsequently relapsed. Patient no. 1508 had no PCR-detectable MRD in PB or BM before relapse, although the original malignant clone containing the identical BCL-1/IgH rearrangement was detected at the time of clinical relapse on lymph node biopsy. Six patients who had undergone ABMT and the two patients who received sibling donor BM remained in continuous CR from 12 to 37 months after BMT. Two of these patients (no. 1698 and 1917) have persistence of MRD after ABMT, whereas four patients have no detectable MRD and have not relapsed after ABMT. Of note, no PCR-detectable disease was found in the allogeneic and syngeneic transplanted patients.
The purpose of this study was to determine whether or not there was evidence of MRD as assessed by PCR analysis of PB or BM samples at the time of and after BMT. Because of the small sample size, albeit the largest reported study of MCL patients, and the rarity of these patients becoming PCR negative, it was not possible to assess the relative risk of relapse associated with persistence of PCR after purging or after BMT. In contrast to our previous studies in follicular lymphoma in whom detection of MRD appeared to be more sensitive when using BM compared with PB,24 in the paired BM and PB samples available at follow-up from these patients we found only one instance in which BM contained PCR-detectable disease, but PB obtained at the same time did not. This suggested that PB may be as useful as BM to follow MRD in these patients, although further studies will be required to determine if this is the case. Of note, in the six patients in whom there was both a PCR amplifiable BCL-1/IgH and an IgH rearrangement, identical results were obtained using both methods on all samples analyzed at the time of and after BMT, despite the fact that the methodology for PCR amplification of the BCL-1/IgH translocation is more sensitive. This suggests that there were no patients in the present study who had PCR-detectable disease below the limit of detection using the ASO primers.
DISCUSSION
The aim of this present study was to assess the clinical significance of PCR-detectable MRD in patients with MCL undergoing BMT. Although several investigators have described PCR amplification of the BCL-1/IgH translocation occurring in MCL,15,16,18,36,37 none have used it to assess the clinical significance of detection of MRD in serial samples. To our knowledge only one MCL patient, who was undergoing allogeneic BMT, has been studied for MRD detection after therapy using PCR amplification of the clonal rearranged IgH gene.25 We showed that semi-nested PCR amplification of the BCL-1/IgH translocation and the clonal rearranged IgH gene are feasible and sensitive methods to detect and follow MRD in 19 of 26 (73%) patients with MCL. The two molecular markers provided a limit of detection of one residual lymphoma cell in 103 to 105 normal cells. In addition, the CDR III sequence acquired from relapse samples were 100% aligned to the CDR III sequence from the diagnostic samples confirming that the CDR III region is stable in MCL and that the sequence obtained represents the malignant clone.
Although the majority of patients with MCL exhibit a t(11; 14) not all cases can be PCR amplified. In this study we used primers spanning the major translocation cluster in the BCL-1 locus and were able to amplify 33% of cases, in keeping with previously published studies.16,17,36,38 To increase the number of patients followed, we sequenced the CDR III regions of the IgH and designed patient specific oligonucleotide primers. This was successful in 61% of the patients studied. Whereas it may be possible to use alternative strategies to sequence and PCR amplify clonal IgH rearrangements, we chose to use the relatively simple strategy using one set of VH family specific primers for PCR amplification. In contrast to our previous findings in follicular lymphoma21 and CLL20 we were not able to increase the yield significantly by employing VH consensus primers. The reason for this in the patients with MCL is unclear because the sequence of this primer was identified in the IgH VH regions sequenced using the VH family specific primers, but this may be a result of a somatic mutation arising within the germline configuration in these patients.
Unlike previously published studies,30,31 use of the VH4 family appeared relatively over-represented in our patients with MCL. Additionally, and also in contrast to these previously described studies,30,31 we show that replacement mutations occur significantly in excess in the CDR I and CDR II regions of the rearranged IgH gene in MCL (P = .05). These findings indicate, as described in other B-cell malignancies,28,29,39 that the final malignant transformation in MCL is likely to happen in a mature B cell that has been stimulated by antigen. This feature was not seen in five of the patients studied, but these clones might express polyreactive antibodies not dependent on somatic mutation for ongoing stimulation by antigens leading to final transformation.40 41
Assessment of the clinical significance of MRD after high-dose therapy showed that of the 10 patients in whom PCR-detectable MRD persisted, 8 patients relapsed. This is similar to our findings in follicular lymphoma and B-cell CLL.20,21,32 42 The findings suggest that our therapeutic goal should be to eradicate all PCR-detectable MRD in these patients and also suggest that additional treatment is necessary for cure. Of the six patients with no PCR detectable MRD after BMT, one patient relapsed. This patient had BM infiltration with the original clone at time of relapse. Whether the malignant clone arose in the BM from levels that were undetectable by PCR at the time of the follow-up before clinical relapse, or the lymph node was the site of relapse and then tumor cells spread secondly to the BM is unknown. Therefore, although our results suggest that persistence or reappearance of PCR-detectable MRD suggests poor prognosis, further studies with larger patient numbers and longer follow-up will be required to determine whether eradication of PCR-detectable MRD is associated with cure of disease.
The poor results obtained by immunological purging with MCL are in contrast to our results obtained in other B-cell malignancies in which purging was successful in 50% of the cases.20,23 When comparing the efficacy of immunological purging in MCL and B-cell CLL, a significant difference was detected in the two disease settings. The tumor load of the harvested BM products in MCL and CLL appeared to be similar because all patients had less than 10% infiltration. MCL cells remaining after purging might represent a population of cells that do not express, or express only weakly, the target antigens. However, MCL cells have high expression of CD20,17 which is included in the MoAb cocktail. Although MCL cells are known to be CD10 negative, anti-CD10 MoAb was included in the cocktail because clonogenic MCL cells might express earlier lineage antigens than those seen in the more mature lymphoma cells in the lymph node. Unfortunately, in the present study there was no advantage in adding anti-CD19 MoAb to the cocktail used for purging. It is unclear whether resistance to purging is caused by immature B cells that contain clonogenic potential but do not express the target antigens. However, our studies of somatic mutations in MCL suggest that the final transformation occurs in a mature B cell. Alternatively, MCL cells might simply be resistant to complement mediated lysis. We previously showed that follicular lymphoma cells that failed to be purged by complement mediated lysis could be eradicated successfully by purging with immunomagnetic beads.43 Studies to assess this possibility in MCL are presently underway.
Reinfusion of tumor cells in autologous BMT has been shown to contribute to relapse in gene-marking studies.44,45 Additionally, we have shown an association between eradication of MRD from the harvested BM before reinfusion and improved disease-free survival in other B-cell malignancies.20 23 Our findings here indicate that high-dose therapy and ABMT with immunological purging by complement mediated lysis does not improve the unfavorable outcome in MCL. The finding that the vast majority of patients were reinfused with BM that contained residual lymphoma cells and that the outcome of ABMT was so poor in this disease is disturbing. Whereas it remains possible that MCL cells are intrinsically resistant to high-dose chemoradiotherapy, these results are also consistent with the notion that infusion of lymphoma cells at ABMT might be associated with subsequent relapse. Future studies at this institute will be directed towards developing techniques by positive selection of stem cells and/or by negative depletion of MCL cells to obtain a source of stem cells for rescue after high-dose therapy that are free of tumor contamination. These preclinical studies using different immunological purging and positive selection procedures will be evaluated using the methods described here with the aim being to eradicate PCR-detectable disease from the source of autologous stem cells.
Supported by Grant No. CA66996 (to J.G.G.) from the National Institutes of Health, Bethesda, MD. N.S.A. was supported by an American Traveling Fellowship funded by The Finsen Center, Rigshospitalet, University Hospital of Copenhagen and The Danish Medical Research Council, Denmark.
Address reprint requests to John G. Gribben, MD, PhD, Division of Hematologic Malignancies, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115.