Sixty-four patients with adult T-cell leukemia/lymphoma (ATL; 18 patients with indolent subtype and 46 with aggressive subtype) associated with human T-lymphotropic virus type 1 (HTLV-1) were analyzed using comparative genomic hybridization (CGH). The most frequent observations were gains at chromosomes 14q, 7q, and 3p and losses at chromosomes 6q and 13q. Chromosome imbalances, losses, and gains were more frequently observed in aggressive ATL than in indolent ATL, with significant differences between the 2 ATL subtypes at gains of 1q and 4q. An increased number of chromosomal imbalances was associated with a significantly shorter survival in all patients. A high number of chromosomal losses was associated with a poor prognosis in indolent ATL, whereas the presence of 7q+ was marginally associated with a good prognosis in aggressive ATL. Paired samples (ie, samples obtained at different sites from 4 patients) and sequential samples from 13 patients (from 6 during both chronic disease and acute crisis and from 7 during both acute onset and relapse) were examined by CGH and Southern blotting for HTLV-1. All but 2 paired samples showed differences on CGH assessment. Two chronic/crisis samples showed distinct results regarding both CGH and HTLV-1 integration sites, indicating clonal changes in ATL at crisis. In 11 patients, the finding of identical HTLV-1 sites and clonally related CGH results suggested a common origin of sequential samples. In contrast to chronic/crisis samples, CGH results with all acute/relapse sample pairs showed the presence of clonally related but not evolutional subclones at relapse, thereby suggesting marked chromosomal instability. In summary, clonal diversity is common during progression of ATL, and CGH alterations are associated with clinical course.
Introduction
Adult T-cell leukemia/lymphoma (ATL), a distinct clinicopathological disorder, is a peripheral T-lymphocytic malignant disease caused by human T-lymphotropic virus type 1 (HTLV-1).1,2 The diverse clinical features of this disease led to its subclassification into acute, lymphoma, chronic, and smoldering subtypes. The median survival time (MST) is 6.2 months, 10.2 months, and 24.3 months, respectively, for the first 3 forms3; an MST for the smoldering subtype has not been established. A multivariate analysis found that advanced performance status (PS), a high level of lactic acid dehydrogenase (LDH), age above 40 years, a large number of lesions, and hypercalcemia are the principal factors associated with shorter survival in all forms of ATL.4 Several other prognostic factors have been identified, such as Ki 67 expression in the ATL cells, immunophenotypes other than the typical CD4+CD8− ATL cells, defective HTLV-1 integration, and deletion of the p15INK4B or p16INK4A gene.5-8
The incidence of ATL is relatively low among carriers of HTLV-1, and those in whom the disease develops usually have a 40- to 70-year latency period from the time of infection to progression to ATL.9 A statistical analysis suggested that development of ATL requires 5 independent genetic events.10 The tumor suppressor genes p53, p15INK4B, and p16INK4A are altered in aggressive ATL.8,11 Sometimes these abnormalities appear at the time of crisis. Chromosome abnormalities in ATL, including numerical and structural aberrations, occur more often in aggressive ATL than in indolent ATL.12 No cytogenetic abnormality specific to ATL has been found.12 Overall, multistep leukemogenesis in ATL remains largely enigmatic.13
Chromosomal banding techniques are useful for identifying chromosomal aberrations and imbalances but are less helpful for recognizing potentially amplified regions. Furthermore, this technique requires analysis of metaphases after cell culture, which is time-consuming and may select subclones that had a growth advantage in vitro. Comparative genomic hybridization (CGH) allows rapid analysis of chromosomal imbalances without the requirement of cell culturing and is more reliable than cytogenetic studies for recognizing potentially amplified regions.14 The aims of this study were to determine the chromosomal imbalances that may play a role in the development and progression of ATL and to determine the clinical relevance of these genetic alterations.
Materials and methods
Patients
We assessed 64 Japanese patients with ATL, 18 of whom had the indolent type (all chronic type) of the disease and 46 of whom had the aggressive type (41 acute type and 5 lymphoma type). In all cases, the diagnosis of ATL was based on clinical features, immunophenotypes, the presence of anti–HTLV-1 antibody, and monoclonal integration of HTLV-1 proviral DNA. The subtypes of ATL were defined as described previously.3 Briefly, aggressive ATL (acute or lymphoma type), in contrast to indolent ATL (chronic or smoldering type), includes organ involvement, such as that characterized by lymphadenopathy, hypercalcemia, or a high LDH (more than twice the upper limit of the normal range). The last follow-up evaluations in this study were done in May 2000.
Anti–HTLV-1 antibody, immunophenotypes, and HTLV-1 proviral DNA integration
Serum samples were examined for antibody to HTLV-1 by using immunofluorescence assays or enzyme immunoassays.15,16 The surface phenotypes of ATL cells were determined with a panel of monoclonal antibodies by using an immunofluorescence assay or a Spectrum III apparatus (Ortho Diagnostic System, Raritan, NJ) as described previously.6 Monoclonal integration of HTLV-1 proviral DNA into ATL cells was assayed as described previously.7
CGH
CGH experiments were performed as described previously, with minor modifications.17 18 Normal human genomic DNA (reference DNA) and tumor DNA were labeled by standard nick-translation procedures with digoxigenin-11-deoxyuridine triphosphate (dUTP) and biotin-16-dUTP (Roche Diagnostics, Mannheim, Germany), respectively. Labeled reference DNA and tumor DNA (500 ng each) were precipitated in the presence of Cot-1 DNA (Life Technologies, Eggenstein, Germany) and salmon-sperm DNA (Sigma, Heidelberg, Germany) and hybridized to metaphase spreads prepared from blood from a healthy male donor. After 3 to 4 days of hybridization, digoxigenin- and biotin-labeled DNAs were detected by using CY-3 and fluorescein isothiocyanate, respectively. Chromosomes were counterstained with 4,6-diamidino-2-phenylindole (DAPI), resulting in a G-banding–like pattern that was used for chromosome identification. Image acquisition, processing, and evaluation were done with a Leica DM RXA epifluorescence microscope equipped with a Sensys CCD camera (Kodak KAF 1400 chip; Photometrics, Tucson, AZ) controlled by Leica Q-FISH software (Leica Microsystems Imaging Solutions, Cambridge, United Kingdom). Three-color images—green for tumor DNA, red for reference DNA, and blue for the DAPI counterstaining—were acquired from 12 to 18 metaphases per sample. Images were processed by using Leica Q-CGH software. The threshold values for detection of genomic imbalances were determined as 0.75 for losses and 1.25 for gains.
Statistical analysis
Survival in the patients with indolent ATL and in those with aggressive ATL was calculated according to the Kaplan-Meier method and compared by using the log-rank test. The incidence of clinical features among subgroups was analyzed with the Pearson χ2 test or the Mann-Whitney U test.
Results
CGH findings
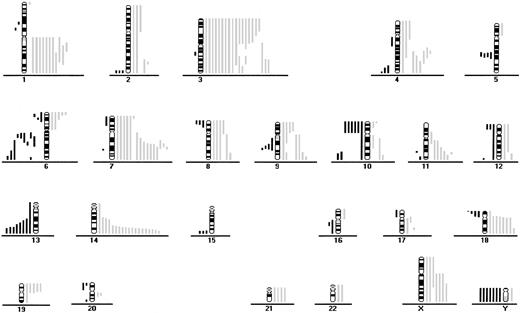
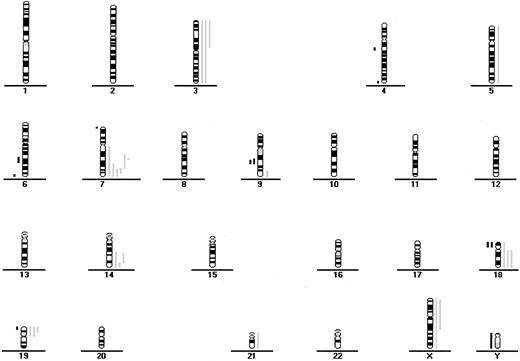
Fifty-six of the 64 patients (88%) had gains or losses of genetic material (Figure 1 and Figure2). Among samples from all patients with ATL, the most frequent chromosomal gains were at 14q (34%), 7q (33%), and 3p (31%), with the commonly overrepresented region at 7q21-35, 14q32-qter, and 3p21-pter. The most frequent chromosomal losses occurred at 6q (17%), with 3 delimited consensus regions at 6q14-q16, 6q21-q22 and 6q27-qter, and 13q (13%) with the minimum common region at 13q32-qter (Table 1 and Table2 and Figure 1 and 2).
CGH data from 46 patients with aggressive ATL.
Loss of chromosomal material is indicated on the left side and gain of chromosomal material on the right. Each line indicates the region gained or lost in a single tumor.
CGH data from 46 patients with aggressive ATL.
Loss of chromosomal material is indicated on the left side and gain of chromosomal material on the right. Each line indicates the region gained or lost in a single tumor.
CGH data from 18 patients with indolent ATL.
Each line indicates the region gained (right side) or lost (left side) in a single tumor.
CGH data from 18 patients with indolent ATL.
Each line indicates the region gained (right side) or lost (left side) in a single tumor.
Chromosome imbalances (P = .0006), losses (P = .0049), and gains (P = .001) were significantly associated with aggressive ATL compared with indolent ATL (Table 3 and Figures 1 and 2). With respect to individual alterations, there were significantly more gains at 1q and 4q in aggressive ATL than in indolent ATL (Table 1). Gains at 3q and 11q and losses at 13q were marginally significantly different in the 2 ATL subtypes (Tables 1 and 2). The frequency of gains at 7q was similar in indolent and aggressive ATL (33% versus 33%).
Clinical importance of CGH imbalance
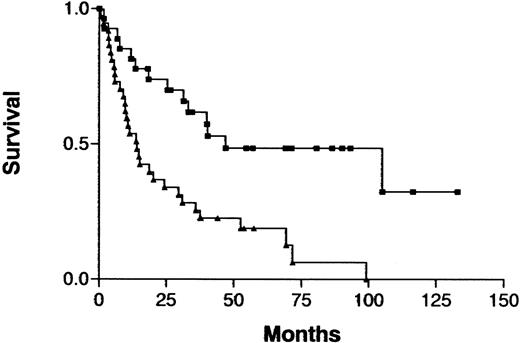
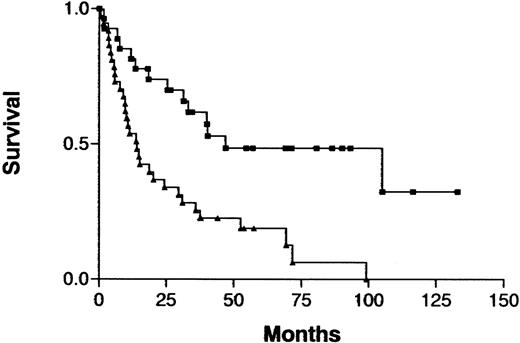
The clinical characteristics in this series of patients with ATL were similar to those previously reported: median age, 58 years (range, 31-80 years); predominantly male (male:female ratio, 2.8:1); and indolent (chronic type) ATL versus aggressive (acute or lymphoma type) ATL was 18 versus 46 cases.3,19 An increased number of imbalances (0-2 versus > 2) on CGH analysis was associated with a significantly shorter survival in the total cohort of patients (P = .0005; Figure 3) but not when either those with indolent ATL or those with aggressive ATL were analyzed separately. Analysis of prognostic factors showed that patients with an increased number of imbalances more frequently had poor prognostic factors, such as advanced PS or a high LDH value, and more aggressive clinical subtypes than other patients (Table4). Patients with an increased number of imbalances had ATL cells with an unusual immunophenotype more frequently than did other patients, although the difference was not significant. Patients with aggressive ATL were treated with doxorubicin, vincristine, cyclophosphamide, and one or several other antineoplastic agents.19 Patients with indolent ATL were either followed without therapy or received a moderate treatment regimen (eg, prednisolone or cyclophosphamide alone) until development of acute crisis of the disease.19
Survival curves for patients with ATL, according to the number of chromosomal imbalances.
Imbalances 0-2 (▪) versus imbalances > 2 (▴) per patient;P = .0005
Survival curves for patients with ATL, according to the number of chromosomal imbalances.
Imbalances 0-2 (▪) versus imbalances > 2 (▴) per patient;P = .0005
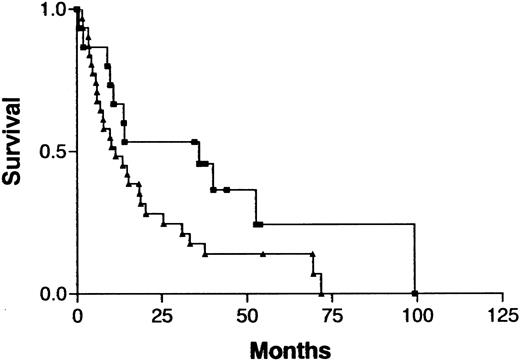
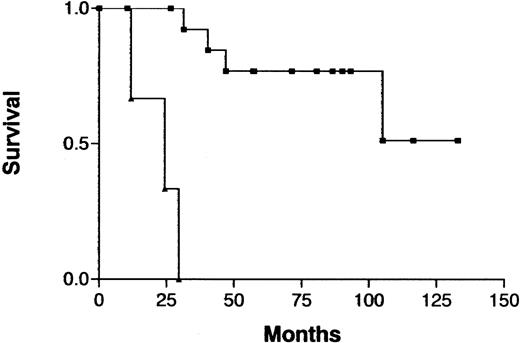
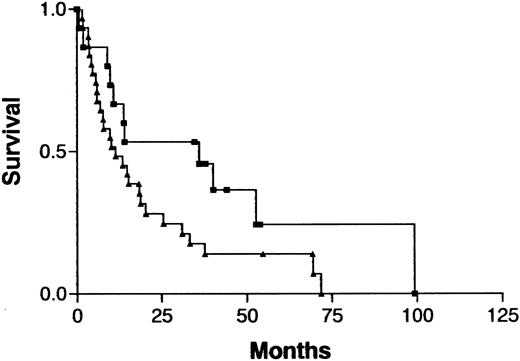
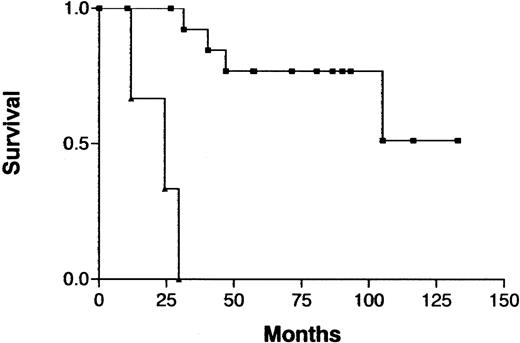
Because treatment and prognosis (MST in this study for aggressive and indolent ATL, 426 days and 3197 days, respectively;P < .0001) were extremely different in patients with indolent ATL compared with those with aggressive ATL, we investigated a possible association between CGH alterations (number of gains, losses, and imbalances and the presence or absence of individual alterations) and prognosis for each group separately. Among patients with aggressive ATL, none of the CGH alterations were significantly associated with prognosis. Interestingly, however, 7q+ was marginally associated with a better prognosis in this cohort (Figure4; P =.069). Furthermore, patients with aggressive ATL and 7q+ and 14q+ had a significantly better prognosis than other patients with aggressive ATL (MST, 1596 days versus 409 days; P =.023). An increased number of chromosomal losses (0-1 versus >1) was the only factor associated with poor prognosis (P < .0001) in patients with indolent ATL (Figure 5). High serum levels of both LDH and blood urea nitrogen or a low serum albumin level, all of which were previously reported to be poor prognostic factors in chronic ATL,3 were observed in 2 of 3 patients with indolent ATL and an increased number of chromosomal losses and in 4 of 15 patients with indolent ATL and fewer chromosomal losses. The locus in the 3 patients with an increased number of chromosomal losses consisted of 6q21-q22 and 9q21-q22; 7p22 and 19p13.3; and 4q21, 6q27-qter, 9q22, and whole 18p; respectively.
Survival curves for patients with aggressive ATL, according to gain at 7q as identified by CGH analysis.
Gain at 7q (▪) versus normal 7q (▴); P = .069.
Survival curves for patients with aggressive ATL, according to gain at 7q as identified by CGH analysis.
Gain at 7q (▪) versus normal 7q (▴); P = .069.
Survival curves for patients with indolent ATL, according to the number of chromosomal losses.
Chromosomal losses of 0-1 (▪) versus > 1 (▴) loss per patient;P < .0001.
Survival curves for patients with indolent ATL, according to the number of chromosomal losses.
Chromosomal losses of 0-1 (▪) versus > 1 (▴) loss per patient;P < .0001.
Clonal analysis by CGH studies and HTLV-1 proviral integration of samples collected either from different sites or sequentially
Samples from different sites (peripheral blood and lymph node in 3 patients and skin and ascites in one patient) were examined by both CGH analysis and Southern blotting for HTLV-1 integration at the onset of acute disease (Table 5). Each pair of samples showed identical HTLV-1 integration sites, indicating that the 2 samples had the same clonal origin. However, in 2 of the 4 sample pairs (those from patients 29, 38, 43, and 49), additional CGH imbalances were present in one of the 2 samples, indicating clonal evolution. The other 2 sample pairs had an identical imbalance (+3p and −13q in patient 38 and −Y in patients 43) in the 2 samples. However, other imbalances were unique, indicating the presence of clonally related but not evolutional subclones at different sites.
We sequentially examined CGH and HTLV-1 integration in 13 patients with ATL. Six matched samples were obtained from patients 11, 23, 57, 62, 63, and 64 during the indolent and acute-crisis phase of their disease, and 7 matched samples were obtained from patients with aggressive ATL initially (patients 2, 6, 16, 20, 27, 30, and 58) and at the time of relapse after chemotherapy and remission. The median duration of follow-up was 893 days (range, 230-2220 days) for the indolent/crisis sample pairs and 497 days (range, 149-3395 days) for the aggressive/relapse sample pairs. Two indolent/crisis sample pairs (those from patients 23 and 62) showed distinct CGH and HTLV-1 integration results, indicating a clonal change in ATL. The other 4 matched indolent/crisis sample pairs showed an identical HTLV-1 integration site and either no or some additional alterations in CGH at crisis, consistent with clonal evolution in the latter samples. In contrast to the chronic/crisis sample pairs, all the acute/relapse sample pairs showed an identical HTLV-1 integration site, indicating that relapse originated from the same ATL clones as those in the acute phase. In all matched acute/relapse sample pairs, at least one imbalance in the acute phase was unique, indicating the presence of clonally related but not evolutional subclones at relapse, as in the samples from different sites from patients 38 and 43.
Sequential examinations in 6 patients (2 indolent/crisis matched sample pairs and 4 aggressive/relapse matched sample pairs) showed 7q+. In 5 of the 6 patients, the abnormality was in both samples; the sixth patient had 7q+ in the initial sample only. Thus, 7q+ is apparently not associated with disease progression.
Discussion
In this study, we identified an elevated number of chromosomal imbalances in ATL. Previous cytogenetic analyses of ATL identified gains at chromosomes 3, 7, and 21 and losses at Y, 6q, 10p, 3q, 5q, 9q, 13q, 1p, and 7p.12 Our results confirm the idea that these chromosomes are recurrent targets of alterations in ATL, but the frequency of each alteration was higher on CGH analysis than on conventional cytogenetic studies. We also observed certain imbalances, such as gains at 14q32 and 7q21-q35, not previously recognized by classic cytogenetic assessment. A recent, smaller CGH study of ATL using 8 cell lines and 12 primary samples also showed frequent gains at 7q and 14q.20 However, that study contained a significant number of primary samples (5 of 12 acute ATL samples) without chromosomal aberrations and a high frequency (6 of 20, or 30%) of gains at 2p. All gains at 2p were restricted to cell lines, suggesting that this change occurred in vitro.
Gains at 14q32 and 7q21-q35 are particularly frequent in ATL. A gain at 14q32 appears to be fairly specific for ATL, although a few reports have noted its occurrence in other malignant diseases.21-23 The TCL1 oncogene lies at 14q32 and, in T-prolymphocytic leukemia, is often involved in chromosomal translocation with one of the T-cell receptor (TCR) loci.24 However, 2 studies showed that TCL1 is not amplified in ATL samples with 14q32+, suggesting that another oncogene on 14q32 might be responsible for the multistep leukemogenesis in ATL.20,25 Gain at 7q21-q35 is frequently observed in glioblastoma, renal cancer cell lines, choriocarcinoma, a drug-selected colon cancer cell line, and drug-refractory acute lymphoblastic leukemia.26-28 Multidrug resistance gene 1 (MDR1) lies at 7q21.1 and is amplified in a colon cell line having a gain at 7q21-q35. Thus, MDR1 is one of the candidate genes at 7q21-q35 responsible for ATL leukemogenesis. Overexpression of MDR1 is detected in many drug-resistant malignant diseases, including ATL.29,30 We also found that losses at 6q and 13q32-qter are particularly frequent in ATL. Loss at 6q and 13q32-qter is observed in many malignant diseases, including lymphoid hematologic cancers.31,32 No altered tumor suppressor genes at these 2 loci have thus far been identified.32
Clinical subclassification of ATL is useful for predicting prognosis and guiding therapeutic decisions.33 In the current study, we demonstrated that patients with aggressive ATL have a higher number of chromosomal imbalances, losses, and gains than those with indolent ATL. A similar observation has been reported for other types of malignant diseases as they evolve from low- to high-grade status.34 35 In addition, we found that specific alterations, including gain at 1q and 4q and loss at 13q are more frequent in aggressive ATL, suggesting that these alterations could be important in progression of ATL.
Several genetic alterations, such as defective HTLV-1 integration and deletion of the p15INK4B or p16INK4A gene, were reported to be prognostic factors in ATL.7,8 However, no previous study analyzed the clinical importance of whole chromosomal alterations in ATL by using either cytogenetic analysis, allelotyping, or CGH. In this study, we examined the clinical importance of CGH alterations in ATL. An increased number of CGH imbalances was significantly associated with a shorter survival in the entire group of patients but not when patients were evaluated according to clinical disease subtype. The 7q+ abnormality was marginally associated with a good prognosis in aggressive ATL. The frequency of this alteration was similar in indolent and aggressive ATL. Among sequential samples, 5 matched sample pairs showed a gain at 7q in both samples and the sixth had the gain at diagnosis of acute ATL but not at relapse. These observations suggest that a gain at 7q might be important in the earlier stages of the multistep leukemogenesis in ATL. This is the first report of a genetic alteration possibly associated with indolent ATL. The characteristics of chemotherapy-induced clinical remissions in patients with aggressive ATL are heterogeneous.19 The 7q alteration might be responsible for part of the heterogeneity.
CGH analysis of samples from patients with indolent ATL showed that an increased number of chromosomal losses was associated with a poor prognosis. Deletion of the tumor suppressor genes p15INK4B and p16INK4A was previously associated with a poor prognosis in indolent ATL.8 The chromosomal losses in the patients with indolent ATL in this study were diverse and did not include 9p21, where p15INK4B and p16INK4A are located, suggesting that alteration of many tumor suppressor genes is important in the late stage of multistep leukemogenesis in ATL.
Paired samples obtained either from different sites or sequentially were examined by CGH and Southern blotting for HTLV-1 integration. All but 2 paired samples showed some differences on CGH assessment. A clonal change in ATL was identified in 2 of 6 matched chronic/crisis sample pairs on the basis of nonoverlapping CGH alterations and novel HTLV-1 integration results. This confirms the previous observation of clonal change in about 20% of patients with ATL at crisis, as detected by Southern blotting examination for both HTLV-1 integration and TCR-β rearrangement.7 In contrast to the clonal evolution characteristic of multistep carcinogenesis in most human malignant diseases, the frequent clonal change in ATL at crisis is a peculiar phenomenon and probably reflects the emergence of multiple premalignant clones in viral leukemogenesis.7,36,37 The sequential samples from 11 patients showed identical HTLV-1 bands and clonally related CGH results, indicating that these paired samples had the same clonal origin. Nevertheless, a prominent difference in CGH results occurred between the matched chronic/crisis samples and the matched acute/relapse samples. All of the 4 matched chronic/crisis sample pairs with identical HTLV-1 bands had either no or some additional imbalances in CGH at crisis, consistent with clonal evolution. In all of the 7 matched acute/relapse sample pairs, there was at least one unique CGH change at the acute phase, indicating the presence of clonally related but not evolutional subclones at relapse. This phenomenon was observed previously in several aggressive malignant diseases and may reflect chromosomal instability as described in a tumor-progression model.38-40 According to the model, the population of neoplastic cells can evolve toward greater complexity and genetic divergence or the population can become more homogeneous during progression.38 Apparently, the convergence-divergence idea is compatible with the multistep leukemogenesis in ATL. An oligoclonal HTLV-1–infected T-cell population may be detected in asymptomatic HTLV-1 carriers, as previously reported by us41 and others.37 During the indolent phase of disease, the ATL clone is stable on cytogenetic analysis (K.T. et al, unpublished data, September, 2000) and Southern blotting for HTLV-1 and TCR-β.7 At acute crisis emerging from indolent ATL, clonal evolution or clonal change is frequently detected, as it was in this study and others.7 In this study, we found that during the last step of multistep leukemogenesis (ie, acute ATL), clonally related but not evolutional subclones frequently occurred at different sites or at relapse.
In summary, clonal diversity is common during progression of ATL, and CGH alterations are associated with clinical disease course. The mechanism of chromosomal instability in ATL remains unknown but may be associated with defects in genes acting as a cell-cycle checkpoint.42
Supported in part by grants from the US National Institutes of Health and Defense Department, and by the Lymphoma Research Foundation, C. and H. Koeffler Trust, Horn Foundation, Parker Hughes Trust, and the KoSo Foundation. H.P.K. holds the Mark Goodson Endowed Chair of Oncology Research.
K.T. and J.K. contributed equally to this work and should be considered first authors.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kunihiro Tsukasaki, Department of Hematology, Molecular Medicine Unit, Atomic Bomb Disease Institute, Nagasaki University School of Medicine, 1-12-4 Sakamoto, Nagasaki 852, Japan; e-mail: tsukasak@net.nagasaki-u.ac.jp.