Abstract
We recently identified aberrant cytoplasmic expression of nucleophosmin (NPM) as the immunohistochemical marker of a large subgroup of acute myeloid leukemia (AML) (about one-third of adult AML) that is characterized by normal karyotype and mutations occurring at the exon-12 of the NPM gene. In this paper, we have elucidated the molecular mechanism underlying the abnormal cytoplasmic localization of NPM. All 29 AML-associated mutated NPM alleles so far identified encode abnormal proteins which have acquired at the C-terminus a nuclear export signal (NES) motif and lost both tryptophan residues 288 and 290 (or only the residue 290) which determine nucleolar localization. We show for the first time that both alterations are crucial for NPM mutant export from nucleus to cytoplasm. In fact, the cytoplasmic accumulation of NPM is blocked by leptomycin-B and ratjadones, specific exportin-1/Crm1-inhibitors, and by reinsertion of tryptophan residues 288 and 290, which respectively relocate NPM mutants in the nucleoplasm and nucleoli. NPM leukemic mutants in turn recruit the wild-type NPM from nucleoli to nucleoplasm and cytoplasm. These findings indicate that potential therapeutic strategies aimed to retarget NPM to its physiological sites will have to overcome 2 obstacles, the new NES motif and the mutated tryptophan(s) at the NPM mutant C-terminus.
Introduction
In acute myeloid leukemia (AML), a clinically and molecularly heterogeneous disease,1 recurrent cytogenetic abnormalities help define subgroups with different prognosis, and identify patients who might benefit from targeted therapies.1 However, almost half adult AMLs display normal karyotype at conventional cytogenetics,2 and the clinical and molecular features of this large subgroup of patients are still poorly understood.3,7
We recently observed that about 60% of adult AML with normal karyotype display aberrant cytoplasmic expression of nucleophosmin (NPM).8 A multifunctional protein9,15 that characteristically shuttles between the nucleus and the cytoplasm,16 NPM is found mainly in the nucleolus,17,19 where it is one of the most abundant of the approximately 700 proteins so far identified by proteomic techniques.20 Cytoplasmic NPM identifies a distinct subgroup of AML, named NPMc+ AML, that accounts for about 35% of all adult AML and is characterized by wide morphologic spectrum, multilineage involvement, high frequency of FLT3-ITD mutations, absence of CD34, and relatively good response to induction therapy.8 NPMc+ AML also has a distinct gene expression profile21 and carries mutations in exon-12 of the NPM gene8 that serve as predictor of favorable prognosis in AML with normal karyotype,22,24 and as a marker for monitoring of minimal residual disease.25
In spite of the close association between the aberrant cytoplasmic expression of NPM and exon-12 NPM mutations,8 the mechanism underlying cytoplasmic accumulation of NPM in leukemic cells and its interference with wild-type NPM protein remained to be elucidated.
Recently, Nakagawa et al26 asserted that the creation of a nuclear export signal (NES) motif at the C-terminus of NPM leukemic mutants (as consequence of the exon-12 NPM mutations) is alone responsible for cytoplasmic dislocation of NPM. In this paper, we provide evidence that the new NES motif created by the mutational event is not in itself sufficient to cause nuclear export of the NPM leukemic mutants and demonstrate it needs to act in concert with the mutated tryptophan(s) 288 and 290 at the mutant C-terminus. These findings have a relevant biological and potentially therapeutic significance.
Materials and methods
Cells for transfection, primary AML samples, and OCI/AML3 cell line
Transfections were performed on NIH-3T3 murine fibroblasts and human lung carcinoma H1299 cells. Lymphoprep-isolated leukemic cells for biochemical studies were obtained from 3 patients with AML carrying NPM mutation A and 2 patients with AML with wild-type NPM. Bone marrow trephines from 20 patients with AML (10 with NPM mutations and 10 without NPM mutations) were B5-fixed/decalcified and embedded in paraffin, as previously described.8 The 10 NPM-mutated cases included 7 mutations A, 1 mutation B, and 2 mutations D. The human AML cell line OCI/AML3, which carries an exon-12 NPM mutation A,27 was provided by H. Drexler and grown in alpha–modified Eagle medium (MEM) plus 10% fetal bovine serum (FBS), 1% glutamine, and antibiotics.
Plasmid constructs
NPM mutants A, B, C, and D were generated by polymerase chain reaction (PCR) using NPMwt as template; the same forward primer (5′-CGCCACGCTAGCGAAGATTCGATGGAC-3′) with different reverse primers were used for each mutant (mutant A: 5′-CTATTTTCTTAAAGAGACTTCCTCCACTGCCAGACAGAGATCTTGAATAGCCTCTTGG-3′; mutant B:5′-CTATTTTCTTAAAGAGACTTCCTCCACTGCCATGCAGAGATCTTGAAT AGCCTCTTGG; mutant C: 5′-CTATTTTCTTAAAGAGACTTCCTCCACTGCCACGCA GAGATCTTGAATAGCCTCTTGG; and mutant D: 5-CTATTTTCTTAAAGAGACTTCCTCCAC TGCCAGGCAGAGATCTTGAATAGCCTCTTGG). The PCR products of mutants A, B, C, and D were cloned into the pcDNA3.1/NT-GFP-TOPO vector (Invitrogen, Carlsbad, CA). To generate the double-flag–HA tag at the N-terminal of both wild-type NPM and mutant A, PCR was performed with either wild-type or mutant A as template; 5′-CGCCACGCTAGCGAAGATTCGATGGAC and 5′-TCAAGAATTCCAGAAATGAAATAAGACGG were used, respectively, as forward and reverse primers. The PCR product was excised by NheI and EcoRI, and the resulting fragment was subcloned into the PCIN4 vector containing the Flag-HA tag at the N-terminal of the inserted fragment.
Constructs pEGFP-C1-NPM_MUT_A_A290W, pEGFP-C1-NPM_MUT_A_C288W+A290W, and pEGFP-C1-NPM_MUT_A_NO_ 2nd_NES were generated using pEGFP-C1-NPMmA8 as a template, by a set of 2 partially self-complementary primers bearing the desired mutation. The 2 primers were annealed, and the resulting double-stranded DNA sequence bearing BglII-EcoRI–compatible ends was ligated into the pEGFP-C1-NPMmA vector previously digested with the same 2 enzymes, exploiting the localization of the mutated region between these restriction sites. The sequences of primers are the following: NPM_ MUT_A_A290W_FOR: 5′-GATCTCTGTCTGTGGGTGGAGGAAGTCTCTTTAAGAAAATAGG-3′; NPM_MUT_A_A290W_REV: 5′-AATTCCTATTTTCTTAAAGAGACTTCCTCCACCCACAGACAGA-3′; NPM _MUT_A_C288W+A290W_FOR: 5′-GATCTCTGGCTGTGGGTGGAGGAAGTCTCTTTAAGAAAATAGG-3′; NPM_MUT_A_C288W+A290W_ REV: 5′-AATTCCTATTTTCTTAAAGAGACTTCCTCCACCCACAGCCAGA-3′; NPM_MUT_A_NO_2nd_NES_FOR: 5′-GATCTCTGTGGAGCAGGGGAGGAAGGCTCTTTAAGAAAATAGG-3′; and NPM_MUT_A_ NO_2nd_NES_REV: 5′-AATTCCTATTTTCTTAAAGAGCCTTCCTCCCCTGCTCCACAGA-3′.
pEGFP-C1-NPM mutants E, G, and pEGFP-C1-NPM_MUT_ A_NO_1st_NES were generated with the QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), using pEGFP-C1-NPMwt8 (for mutants E and G) and pEGFP-C1-NPMmA (for deletion of the first NES) as a template. The manufacturer's instructions were followed except for transformation of bacteria, which was done with 5 μL PCR products instead of 1 μL. Primers were designed on the following sequences: NPM-_MUT_E: 5′-GATCTCTGGCAGTCTCTTGCCCAAGTCTCTTTAAG-3′; NPM_MUT_G: 5′-GATCTCTGGCAGTGCTTCGCCCAAGTCTCTTTAAG-3′; and NPM_MUT_A_NO_1st_NES: 5′-GCACCAGTTATCTGGAAGAACGGGCAGTTTAGGGGCTGG-3′. Every construct was validated by DNA sequencing.
Antibodies
We previously used anti-NPM monoclonal antibodies17,18 that recognize both wild-type and mutated NPM proteins. To study the NPM mutants without the interference of wild-type NPM, we generated an affinity-purified polyclonal antibody (Sil-C) against the synthetic peptide NHCOCH3-CLAVEEVSLRK-COOH (Inbio-Ltd, Tallin, Estonia) corresponding to NPM mutant A. The specificity of Sil-C for the NPM mutant was confirmed by Western blotting (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article). To assess the subcellular distribution of wild-type NPM without the interference of leukemic NPM mutants, we used a mouse monoclonal antibody (clone FC-61991) that recognizes the wild-type but not mutant NPM proteins (Invitrogen, Carlsbad, CA). The antinucleolin/C23 monoclonal antibody, used as a control, was purchased from DakoCytomation (Glostrup, Denmark).
Inhibition of the Crm1-dependent nuclear export pathway
Expression vectors (5 μg) coding HA-NPM wild-type, enhanced green fluorescent protein (eGFP)–NPMmA, or both were transfected in H1299 cells by the calcium-phosphate precipitation method. After 24 hours, cells were treated with 20 nM leptomycin B (LMB; Sigma, St Louis, MO) for 8 hours or left untreated. Cells were fixed with 4% paraformadehyde for immunofluorescence analysis.
NIH-3T3 cells were transfected using Lipofectamine 2000 (Invitrogen) following manufacturer's instructions. After 24 hours, cells grown on glass coverslips were incubated with 10 μg/mL cycloheximide (Merck Biosciences, Nottingham, United Kingdom) (30 minutes), and 20 ng/mL LMB (Merck Biosciences) (5 hours) or 20 ng/mL ratjadones A and C (Alexis Biochemicals, Carlsbad, CA) (5 hours). For immunofluorescence studies, cells were fixed in 4% paraformaldehyde (10 minutes). For time-course experiments, transfected cells were transferred on an Attofluor chamber (Molecular Probes, Eugene, OR) and observed on a MRC-1024 (BioRad, Cambridge, United Kingdom) confocal apparatus mounted on an Olympus IMT-2 microscope with an SPlan 60 ×/1.4 numeric aperture (NA) oil objective (Olympus, Tokyo, Japan). Images of a single slice of cells were recorded in the basal state and after LMB addition at 60-second intervals using the time-series function of the LaserSharp (BioRad) software. The 488-nm line of an argon laser was used for excitation and a 505-550–nm band-pass filter on the PMT2 was used for collecting the fluorescence emission. The images were processed and analyzed with the public-domain ImageJ software (Rasband WS, Image J; US National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/, 1997-2005). Confocal image analysis was performed as previously described.27
For Western blot analysis of eGFP-NPM mutant A subcellular distribution, NIH-3T3 cells transfected with either pEGFP-C1 or pEGFP-C1-NPMmA were incubated with 20 ng/mL LMB (or vehicle, as control) in absence of cycloheximide for either 3 or 6 hours. Cells were then harvested, washed in phosphate-buffered saline (PBS) and lysed in hypotonic buffer according to the method of Schreiber et al,28 slightly modified. The supernatant was saved as cytoplasmic fraction. The pellet fraction, containing nuclei, was washed with hypotonic buffer, solubilized in hypertonic buffer and boiled in SDS before loading. Equivalent dilutions (representing equal number of cells) of the cytoplasmic and pellet fractions were prepared for Western blot analysis of eGFP-NPM mutant A distribution with an anti-GFP monoclonal antibody (Roche, Indianapolis, IN).
The OCI/AML3 cell line cells were seeded in growth medium (106 cells/mL in 24-well plates) and incubated at 37°C with 5% CO2 for 5 hours. Following overnight incubation with LMB (20 ng/mL), cells were washed in PBS, centrifuged, fixed in B5, and embedded in paraffin for immunostaining. Primary leukemic cells from a NPMc+ patient with AML with a high peripheral blood count were processed in the same way.
Immunostaining procedures
For the Flag-HA constructs, H1299-fixed cells were permeabilized with 0.2% Triton-X 100 (10 minutes) and blocked with 10% antigoat serum (30 minutes). Primary antibody anti-HA (1:1000; Roche) was incubated with cells (1 hour), followed by secondary antibody Alexa 568–conjugated antirat (1:1000; Molecular Probes) for 30 minutes. Nuclei were visualized by DAPI staining. Images were collected using a Nikon Eclipse E400 microscope and a 40 ×/1.3 NA oil objective. Pictures were captured by the Spot Camera coupled with Spot 4.09 imaging software (Diagnostic Instruments, Sterling Heights, MI).
The nuclei of NIH-3T3 cells transfected with pEGFP-C1-NPM mutants and pEGFP-C1-NPMwt were visualized with propidium iodide. pEGFP-C1-NPM mutants transfected NIH-3T3 were immunostained with a specific antinucleolin antibody followed by a secondary Texas-Red–conjugated goat antimouse antibody (Southern Biotechnology Associates, Birmingham, AL); nuclei were counterstained with TO-PRO-3 (Molecular Probes). Paraffin-embedded samples from the OCI/AML3 cell line and 1 patient with NPMc+ AML were immunostained with anti-NPM monoclonal antibody.8,29 The primary antibody was revealed using the immunoalkaline phosphatase APAAP technique8,29 and hematoxylin counterstain or a secondary Alexa 488–conjugated goat antimouse antibody (Molecular Probes) and propidium iodide counterstain.
Cellular extracts, Western blotting, and coprecipitation studies
Nuclear and cytoplasmic extracts were prepared either using the NE-PER Nuclear and Cytoplasmic Extraction kit (PIERCE Biotechnology, Rockford, IL) or according to the Schreiber's method,28 slightly modified. For immunoprecipitation, cells were lysed in 1 mL ice-cold lysis buffer (0.5% NP-40, 150 mM NaCl, 25 mM Tris [pH 7.5], 1 mM EDTA, 1 mM Na3VO4, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 1 mM phenylmethylsulfonyl fluoride). After incubation on ice (20 minutes), lysates were centrifuged at 14 000g (10 minutes at 4°C) and then incubated with 4 μg of either control immunoglobulin G (IgG), rabbit anti-NPMm (Sil-C), or mouse anti-NPM (clone 376),8 followed by 30 μL Protein A/G Plus–agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) rocking overnight at 4°C. Beads were extensively washed with buffer containing 0.1% NP-40, 150 mM NaCl, 25 mM Tris (pH 7.5), 1 mM EDTA, and inhibitors. Proteins were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene difluoride (PVDF; Millipore, Billerica, MA), and probed with either rabbit polyclonal anti-Crm1 (Santa Cruz Biotechnology) or mouse monoclonal anti-Crm1 antibody (BD Transduction Laboratories, Palo Alto, CA), followed by horseradish peroxidase–conjugated secondary antibodies. Polypeptides were detected using the enhanced chemiluminescence (ECL) method (Amersham Bioscience, Uppsala, Sweden).
Results
All NPM exon-12 mutations create a leucine-rich NES motif in the carboxy-terminal portion of the NPM protein
Analysis of the 6 NPM mutant forms (A to F) originally identified in NPMc+ AML (Table 1)8 revealed a putative nuclear export signal (NES) motif26 in the C-terminal portion of NPM proteins, which was suggested to be responsible for abnormal NPM cytoplasmic expression. This hypothesis implies that a NES motif is present in all NPM mutant proteins generated by NPM exon-12 mutations.
Changes in tryptophans 288 and 290 and creation of a NES motif in NPM mutant proteins generated by 29 NPM exon-12 mutations in 1554 patients with AML
Mutation . | Nucleotide sequence . | . | . | . | . | Protein sequence . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT* | gaccaagaggctattcaagatct | ctg | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLWQWRKSL | ||||
| A | gaccaagaggctattcaagatct | ctgTCTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| B | gaccaagaggctattcaagatct | ctgCATG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCMAVEEVSLRK | ||||
| C | gaccaagaggctattcaagatct | ctgCGTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCVAVEEVSLRK | ||||
| D | gaccaagaggctattcaagatct | ctgCCTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| E | gaccaagaggctattcaagatct | ctg | gcag | t CTCTTGCCC | aagtctctttaagaaaatag | 286-DLWQSLAQVSLRK | ||||
| F | gaccaagaggctattcaagatct | ctg | gcag | t CCCTGGAGA | aagtctctttaagaaaatag | 286-DLWQSLEKVSLRK | ||||
| E• | gaccaagaggctattcaagatct | ctg | gcag | t CCCTCGCCC | aagtctctttaagaaaatag | 286-DLWQSLAQVSLRK | ||||
| G• | gaccaagaggctattcaagatct | ctg | gcag | t GCTTCGCCC | aagtctctttaagaaaatag | 286-DLWQCFAQVSLRK | ||||
| H• | gaccaagaggctattcaagatct | ctg | gcag | t GTTTTTCAA | aagtctctttaagaaaatag | 286-DLWQCFSKVSLRK | ||||
| Gm | gaccaagaggctattcaagatct | ctgCAGG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCRAVEEVSLRK | ||||
| Km | gaccaagaggctattcaagatct | ctgCCGG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCRAVEEVSLRK | ||||
| Lm | gaccaagaggctattcaagatct | ctgCCGCGGag | t | ggaggaagtctctttaagaaaatag | 286-DLCRGVEEVSLRK | |||||
| Nm | gaccaagaggctattcaagatct | ctgCCAG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCQAVEEVSLRK | ||||
| Om | gaccaagaggctattcaagatct | ctgTTTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| Qm | gaccaagaggctattcaagatct | ctgTCGG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCRAVEEVSLRK | ||||
| 1 | gaccaagaggctattcaagatct | ctg | gcagTCCA | t | ggaggaagtctctttaagaaaatag | 286-DLWQSMEEVSLRK | ||||
| 3 | gaccaagaggctattcaagatct | ctgTCAT | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCHAVEEVSLRK | ||||
| 4 | gaccaagaggctattcaagatct | ctgCTTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| 6 | gaccaagaggctattcaagatct | ctg | gca AGATTTCTTAAATC | gtctctttaagaaaatag | 286-DLWQDFLNRLFKKIV | |||||
| 7 | gaccaagaggctattcaagatctATGC | ctg | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| 12 | gaccaagaggctattcaagatct | ctgGCCC | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCAAVEEVSLRK | ||||
| 13 | gaccaagaggctattcaagatct | ctgTAAG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCKAVEEVSLRK | ||||
| 10 | gaccaagaggctattcaagatct | ctg | gcag | tgCTGCTCCC | aagtctctttaagaaaatag | 286-DLWQCCSQVSLRK | ||||
| 14 | gaccaagaggctattcaagatct | ctg | gcag | t TATTTTCCC | aagtctctttaagaaaatag | 286-DLWQCCSQVSLRK | ||||
| G† | gaccaagaggctattcaagatct | ctgTTTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| H† | gaccaagaggctattcaagatct | ctgCTTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| I† | gaccaagaggctattcaagatct | ctgTAAG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCKAVEEVSLRK | ||||
| J† | gaccaagaggctattcaagatct | ctgTATG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCMAVEEVSLRK | ||||
| IΔ | gaccaagaggctattcaagatct | ctgCAGA | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCRAVEEVSLRK | ||||
Mutation . | Nucleotide sequence . | . | . | . | . | Protein sequence . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT* | gaccaagaggctattcaagatct | ctg | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLWQWRKSL | ||||
| A | gaccaagaggctattcaagatct | ctgTCTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| B | gaccaagaggctattcaagatct | ctgCATG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCMAVEEVSLRK | ||||
| C | gaccaagaggctattcaagatct | ctgCGTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCVAVEEVSLRK | ||||
| D | gaccaagaggctattcaagatct | ctgCCTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| E | gaccaagaggctattcaagatct | ctg | gcag | t CTCTTGCCC | aagtctctttaagaaaatag | 286-DLWQSLAQVSLRK | ||||
| F | gaccaagaggctattcaagatct | ctg | gcag | t CCCTGGAGA | aagtctctttaagaaaatag | 286-DLWQSLEKVSLRK | ||||
| E• | gaccaagaggctattcaagatct | ctg | gcag | t CCCTCGCCC | aagtctctttaagaaaatag | 286-DLWQSLAQVSLRK | ||||
| G• | gaccaagaggctattcaagatct | ctg | gcag | t GCTTCGCCC | aagtctctttaagaaaatag | 286-DLWQCFAQVSLRK | ||||
| H• | gaccaagaggctattcaagatct | ctg | gcag | t GTTTTTCAA | aagtctctttaagaaaatag | 286-DLWQCFSKVSLRK | ||||
| Gm | gaccaagaggctattcaagatct | ctgCAGG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCRAVEEVSLRK | ||||
| Km | gaccaagaggctattcaagatct | ctgCCGG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCRAVEEVSLRK | ||||
| Lm | gaccaagaggctattcaagatct | ctgCCGCGGag | t | ggaggaagtctctttaagaaaatag | 286-DLCRGVEEVSLRK | |||||
| Nm | gaccaagaggctattcaagatct | ctgCCAG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCQAVEEVSLRK | ||||
| Om | gaccaagaggctattcaagatct | ctgTTTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| Qm | gaccaagaggctattcaagatct | ctgTCGG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCRAVEEVSLRK | ||||
| 1 | gaccaagaggctattcaagatct | ctg | gcagTCCA | t | ggaggaagtctctttaagaaaatag | 286-DLWQSMEEVSLRK | ||||
| 3 | gaccaagaggctattcaagatct | ctgTCAT | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCHAVEEVSLRK | ||||
| 4 | gaccaagaggctattcaagatct | ctgCTTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| 6 | gaccaagaggctattcaagatct | ctg | gca AGATTTCTTAAATC | gtctctttaagaaaatag | 286-DLWQDFLNRLFKKIV | |||||
| 7 | gaccaagaggctattcaagatctATGC | ctg | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| 12 | gaccaagaggctattcaagatct | ctgGCCC | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCAAVEEVSLRK | ||||
| 13 | gaccaagaggctattcaagatct | ctgTAAG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCKAVEEVSLRK | ||||
| 10 | gaccaagaggctattcaagatct | ctg | gcag | tgCTGCTCCC | aagtctctttaagaaaatag | 286-DLWQCCSQVSLRK | ||||
| 14 | gaccaagaggctattcaagatct | ctg | gcag | t TATTTTCCC | aagtctctttaagaaaatag | 286-DLWQCCSQVSLRK | ||||
| G† | gaccaagaggctattcaagatct | ctgTTTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| H† | gaccaagaggctattcaagatct | ctgCTTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | ||||
| I† | gaccaagaggctattcaagatct | ctgTAAG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCKAVEEVSLRK | ||||
| J† | gaccaagaggctattcaagatct | ctgTATG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCMAVEEVSLRK | ||||
| IΔ | gaccaagaggctattcaagatct | ctgCAGA | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCRAVEEVSLRK | ||||
Mutations A to F refer to those originally identified in Falini et al.8 Mutations from E• to H• refer to those identified in childhood AML cases in Cazzaniga et al.30 Mutations Gm to Qm refer to those identified in Schnittger et al23 in protocol 99 of the German AML cooperative group; mutations 1, 3, 4, 6, 7, 12, 13, 10 and 14 according to Döhner et al22 ; mutations G†, I†, H† and J† according to Suzuki et al31 ; mutation IΔ according to Verhaak et al.24 Lowercase letters in nucleotide sequences indicate wild-type sequence residues; capital letters, mutated nucleotides. Underlined text indicates leucine-rich NES motif; italics, tryptophan (W) residues.
WT indicates wild type; L, leucin; V, valine; F, phenylalanine; M, methionine; C, cysteine.
NM 002520.
Therefore, we searched for additional exon-12 NPM mutations in patients with AML. Table 1 lists the 29 NPM mutations that have been so far identified by mutational screening of a total of 1554 patients with AML.8,22,24,30,32 In addition to the 6 NPM mutants forms (A to F) we initially reported,8 they include 3 new mutations (G to I) that we identified in 107 pediatric patients,30 6 new mutations (Gm to Qm) that we identified in 401 patients with AML with normal karyotype of the protocol 99 of the German AML Cooperative Group (AMLCG),23 9 new mutations (numbered in Table 1 as 1, 3, 4, 6, 7, 12, 13, 10, and 14) reported by Dohner et al,22 4 new mutations (G† to J†) reported by Suzuki et al,31 and 1 new mutation (Iδ) reported by Verhaak et al.24
We analyzed the NPM mutant proteins derived from the 29 mutation variants listed in Table 1 for the presence of a NES motif, as defined by a short stretch of amino acids characterized by multiple hydrophobic residues with typical spacing (eg, ψXXXψXXψX, where ψ indicates large hydrophobic residues such as leucine, isoleucine, methionine, valine, or phenylalanine).33
The most frequent NES motif was L-xxx-V-xx-V-x-L that was present in 20 (69%) of 29 NPM leukemic mutants. Nine (31%) of the 29 mutants carried a NES motif in which the valine at the second position of NES motif was replaced with another hydrophobic amino acid. In particular, 3 (10.4%) of 29 mutants carried the L-xxx-L-xx-V-x-L motif; 2 (6.9%) of 29 mutants (Table 1; mutations G and H) displayed the L-xxx-F-xx-V-x-L motif; 2 (6.9%) of 29 mutants (Table 1; mutations 10 and 14) had the L-xxx-C-xx-V-x-L motif; and 1 (3.4%) of 29 mutants (Table 1; mutation 1) exhibited the L-xxx-M-xx-V-x-L motif. Of the 29 mutants, 1 (3.4%) (Table 1; mutation 6), generated by a deletion of 7 nucleotides and insertion of 14 nucleotides, showed at the C-terminus the sequence LWQDFLNRLFKKIV. Although this sequence slightly deviates from the accepted NES consensus L-x-(2,3)-(LIVFM)-x(2,3)-L-x-(LI)33 present in the other 29 mutants, it is still consistent with a leucine-rich NES because of the presence of 5 hydrophobic residues (leucine and phenylalanine) within a region of 10 amino acids.34
In conclusion, generation of an additional NES motif in the C-terminal portion of the NPM protein is one common consequence of NPM exon-12 mutations in NPMc+ AML.
Export of NPM leukemic mutants is NES dependent
Finding an additional NES motif suggests cytoplasmic dislocation of NPM may be due to a more active transport of the NPM mutants by Crm1 (exportin 1),35 the receptor for the NES motif of yeast and mammalian proteins.36,37
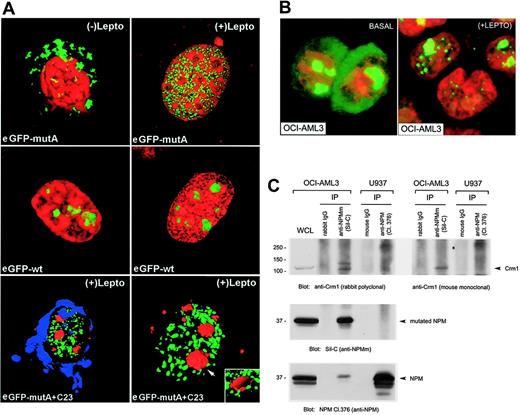
To test this hypothesis, we looked for perturbations in the nucleo-cytoplasmic transport of NPM mutant proteins in the presence/absence of the specific Crm1 inhibitor LMB.35 H1299 or NIH3T3 cells were transfected with plasmids expressing NPM mutants A to D, fused to enhanced green fluorescent protein (eGFP), or NPM wild-type as control, and analyzed by immunofluorescence before and after treatment with LMB. In basal conditions, all 4 NPM mutant proteins were localized in the cytoplasm while NPM wild-type was restricted to the nucleoli, as expected (Figure 1A). However, LMB treatment completely relocated the cytoplasmic NPM mutants A to D into the nucleus (Figure 1B). Identical results (not shown) were obtained in NIH-3T3 cells transfected with mutant A and treated with ratjadones A and C that inhibit specifically the Crm138 with the same mechanism as LMB.39 Time course experiments with mutant A showed that about 50% of protein was relocated in the nucleus at 20 minutes and the process was completed after 1 hour (Figure 1C-D). Western blot analysis of eGFP-NPM mutant A subcellular distribution in LMB-treated NIH-3T3 cells confirmed a time-dependent accumulation of the intact eGFP-NPM mutant A protein (64 kDa) in the pellet fractions (containing the nuclei) (Figure 1E). No alteration of eGFP protein (27 kDa) localization was observed in LMB-treated cells (only a mild contamination of the pellet fractions by the overexpressed protein was detected). In untreated cells, either eGFP or eGFP-NPM mutant A protein were found (except for the contaminant part) only in cytoplasmic fractions, as expected.
Confocal microscopy analysis of eGFP-NPM mutant A (L-xxx-V-xx-V-x-L NES motif) showed that relocalization of the mutated protein induced by LMB was limited to the nucleoplasm (Figure 2A, top panel). This localization clearly differed from that of eGFP-NPM wild-type protein which, as expected, was located in the nucleoli, whether LMB was present or not (Figure 2A, middle panel). Transient transfection with pEGFP-C1-NPMmA and immunostaining for nucleolin/C23 followed by 3D reconstruction confirmed that intranuclear location of NPM mutant A (nucleoplasm, green) and nucleolin (nucleoli, red) were mutually exclusive (Figure 2A, lower panel). A nucleoplasmic staining pattern (not shown) was observed after LMB treatment with 1 rare NPM mutant bearing the NES motif L-xxx-F-xx-V-x-L (mutant G; Table 1).
LMB also caused nuclear relocation of the mutant NPM in the OCI/AML3 cells (Figure 2B) that are known to carry the exon-12 mutation A.27 In these cells, Crm1 and NPM mutant A were shown to coprecipitate, indicating a physical interaction between the 2 molecules (Figure 2C). Leptomycin induced similar nuclear relocation of NPM in primary leukemic cells of 1 patient with NPMc+ AML (not shown).
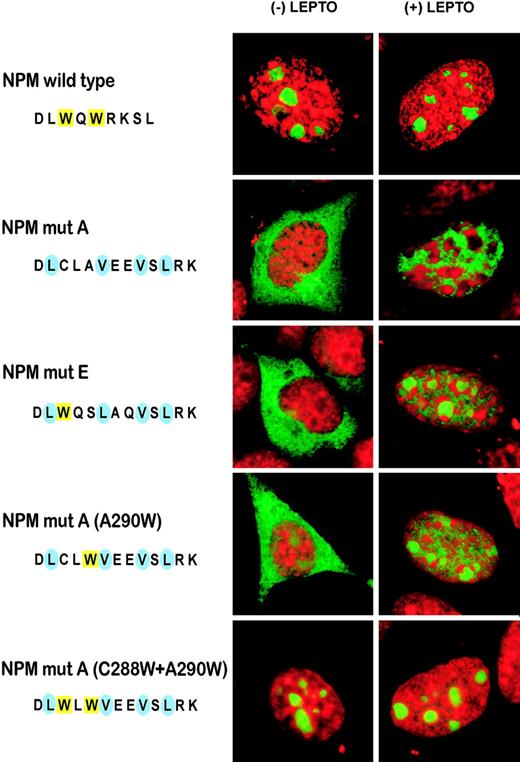
Finally, the requirement of a NES motif for abnormal cytoplasmic expression of NPM was further documented by testing the subcellular localization of a modified eGFP-NPM mutant A construct where the 2 valine residues in the NES motif were replaced by glycines. Transient transfection of NIH-3T3 cells with this construct showed that the NPM mutant accumulated in the nucleoplasm, whether LMB was present or not (Figure 3, right panel).
These results provide evidence that aberrant cytoplasmic localization of NPM mutants in NPMc+ AML is a NES-dependent event.
Two NESs are better than one
The NPM wild-type protein contains a putative physiologic N-terminus NES motif at position 92-104.40 To assess the contribution of the physiologic versus additional C-terminus NES motif created by the mutations in causing abnormal cytoplasmic expression of NPM, we tested the subcellular localization of modified eGFP-NPM mutant A constructs where the second and third hydrophobic residues of the N-terminal or C-terminal NES motif were replaced by glycines (Figure 3). Transient transfection of NIH-3T3 with such constructs showed that the NPM mutants lacking either the physiologic (N-terminus) or the new mutation-related (C-terminus) NES accumulated in the nucleoplasm (Figure 3, middle and right panels), indicating that the nuclear export and cytoplasmic accumulation of NPM mutants can only occur in the presence of 2 NES motifs (Figure 3, left panel); LMB had no effect on subcellular localization of the 2 mutants (not shown).
Nuclear export of NPM mutants is NES dependent. (A-B) H1299 cells transfected with cDNA encoding for the NPMwt and the NPM mutant proteins A to D8 in the absence (A) and presence (B) of LMB. eGFP-NPMwt localizes in the nucleoli both in the presence and absence of LMB. In the absence of LMB, all GFP mutants (A-D) show the expected aberrant cytoplasmic location (A). The same eGFP-mutants are completely relocated in the nucleus in the presence of LMB (B). (C-E) Time-course analysis of LMB induced nuclear retention of eGFP-NPMmutA in NIH-3T3 cells. (C) Confocal microscope images of NIH-3T3 cells at the indicated time points. ○ represents the regions (regions of interest [ROIs]) where the mean fluorescence emission of eGFP-NPMmutA was calculated in the experiment (eg, nucleus [N], cytoplasm [Cy], and the Golgi apparatus [G]). (D) Time-course measurements of mean fluorescence in the selected areas (ROIs) of NIH-3T3 cells. LMB addition produced a loss of fluorescence in the cytoplasm and Golgi apparatus and a concomitant increase in the mean fluorescence of the nucleoplasm. ImageJ software was used for the generation of ROIs and time-course analysis. (E) Western blot analysis of eGFP-NPMmutA subcellular distribution in NIH-3T3 cells with anti-GFP monoclonal antibody. LMB treatment induces a time-dependent accumulation of eGFP-NPMmutA in the pellet fractions (P). Purity of the subcellular fractions was assessed by stripping the membrane and reblotting with an anti–β-tubulin antibody (bottom panel). Only a nonsignificant contamination was detected for the overexpressed proteins (as evident in the GFP blotting) (middle panel). In untreated cells, eGFP-NPMmutA (except for the contaminant part) was found only in the cytoplasmic fractions (Cy). The experiment was conducted in absence of cycloheximide so that continuous presence of GFP-NPMmA in the cytoplasmic fraction during LMB treatment was detected with time.
Nuclear export of NPM mutants is NES dependent. (A-B) H1299 cells transfected with cDNA encoding for the NPMwt and the NPM mutant proteins A to D8 in the absence (A) and presence (B) of LMB. eGFP-NPMwt localizes in the nucleoli both in the presence and absence of LMB. In the absence of LMB, all GFP mutants (A-D) show the expected aberrant cytoplasmic location (A). The same eGFP-mutants are completely relocated in the nucleus in the presence of LMB (B). (C-E) Time-course analysis of LMB induced nuclear retention of eGFP-NPMmutA in NIH-3T3 cells. (C) Confocal microscope images of NIH-3T3 cells at the indicated time points. ○ represents the regions (regions of interest [ROIs]) where the mean fluorescence emission of eGFP-NPMmutA was calculated in the experiment (eg, nucleus [N], cytoplasm [Cy], and the Golgi apparatus [G]). (D) Time-course measurements of mean fluorescence in the selected areas (ROIs) of NIH-3T3 cells. LMB addition produced a loss of fluorescence in the cytoplasm and Golgi apparatus and a concomitant increase in the mean fluorescence of the nucleoplasm. ImageJ software was used for the generation of ROIs and time-course analysis. (E) Western blot analysis of eGFP-NPMmutA subcellular distribution in NIH-3T3 cells with anti-GFP monoclonal antibody. LMB treatment induces a time-dependent accumulation of eGFP-NPMmutA in the pellet fractions (P). Purity of the subcellular fractions was assessed by stripping the membrane and reblotting with an anti–β-tubulin antibody (bottom panel). Only a nonsignificant contamination was detected for the overexpressed proteins (as evident in the GFP blotting) (middle panel). In untreated cells, eGFP-NPMmutA (except for the contaminant part) was found only in the cytoplasmic fractions (Cy). The experiment was conducted in absence of cycloheximide so that continuous presence of GFP-NPMmA in the cytoplasmic fraction during LMB treatment was detected with time.
Role of tryptophans 288 and 290 in nucleolar binding and nuclear export of NPM leukemic mutants
After LMB treatment, NPM mutant A relocated into the nucleoplasm but not in the nucleoli (Figure 2A, top and bottom panels). This finding was not unexpected as both tryptophans 288 and 290 are mutated in this sequence, and the homologous residues in rats (positions 286 and 288) appear to be critical for nucleolar localization of wild-type NPM, with mutations of both tryptophans resulting in nucleoplasmic delocalization of the protein.41
All 29 NPM mutant proteins carried a mutated tryptophan 290, while tryptophan 288 was retained in 9 (31%) of 29 mutants (Table 1). Analysis of tryptophan mutations and NES motif distribution in NPM mutants revealed that the NES most commonly found in the NPM mutants (ie, L-xxx-V-xx-V-x-L) is always associated with mutations of both tryptophans, while tryptophan 288 is retained only in the NPM mutants carrying variants of the most common NES motif (ie, substitution of the valine at the second position of NES with leucine, phenylalanine, cysteine, or methionine).
We used constructs expressing NPM mutant E (Table 1) fused to eGFP to assess the effect of the single mutation at tryptophan 290 on nucleolar binding. After treatment with LMB, cytoplasmic mutant E, like mutant A, relocated to the nucleoplasm; however, mutant E also retained some capability to target nucleoli, as traces of GFP were detected in these structures (Figure 4). A similar pattern was observed using a derivative construct of mutant A with tryptophan 290 reinserted by site-directed mutagenesis (A290W; Figure 4). Notably, when both tryptophans were reinserted in mutant A (C288W + A290W), the mutant protein localized completely to the nucleoli whether LMB was present or not (Figure 4), suggesting a dose effect for tryptophan molecules.
These findings provide evidence that: (1) tryptophan 290 is mutated in all NPM leukemic mutants; (2) tryptophan 288 is retained only in NPM mutants carrying a NES variant other than the most common motif L-xxx-V-xx-V-x-L; (3) as in the rat,41 tryptophans 288 and 290 are both important for nucleoli targeting; (4) maximum dislocation of NPM mutants from nucleoli to nucleoplasm occurs when both tryptophans are mutated; and (5) NES-dependent nuclear export of the NPM mutant A is blocked following reinsertion of both tryptophans 288 and 290.
Identification of the mutant NPM protein by a novel specific antibody
Currently used anti-NPM antibodies17 do not discriminate between wild-type and mutated NPM proteins, which, since NPM exon-12 mutations are heterozygous,8 are both found in leukemic cells.
LMB relocates NPM mutants in the nucleoplasm. (A) Confocal microscopy of NIH-3T3 cells transfected with pEGFP-C1-NPMmA or pEGFP-C1-NPMwt in presence and absence of LMB. In basal conditions, the mutant A is cytoplasmic (top left) but relocates in the nucleoplasm (top right) following incubation with LMB. eGFP-NPMwt localizes in the nucleoli both in the presence and absence of LMB (middle panel, left and right). The bottom panel shows LMB-treated cells transfected with pEGFP-C1-NPMmA (green) and immunostained with an antinucleolin (C23) monoclonal antibody (Texas red)–labeling nucleoli (arrow). The nuclear membrane is blue (TO-PRO-3 staining). After LMB, the eGFP-NPM mutant A was relocated in the nucleoplasm. Nucleolar exclusion was confirmed by an electronic cut of the nucleolar surface (inset in the bottom right figure). (B) The anti-NPM antibody 376 labels both nucleoli and cytoplasm of OCI/AML3 cells in the absence of LMB (left). Incubation with LMB results in nucleoplasmic relocation of the cytoplasmic NPM (right). Images were collected as described27 using a Zeiss Plan Apochromat 100 ×/1.4 NA oil objective. (C) OCI-AML3 and U937 cell lysates were subjected to immunoprecipitation (IP) with either control IgG or rabbit anti-NPMm (Sil-C) and mouse anti-NPM (Cl. 376): Western blots with anti-Crm1 antibodies are shown in the top panels. Blotting with the anti-NPMm (Sil-C) and anti-NPM (Cl. 376) antibody are also shown (middle and bottom panels). WCL indicates OCI-AML3 whole-cell lysate included as control.
LMB relocates NPM mutants in the nucleoplasm. (A) Confocal microscopy of NIH-3T3 cells transfected with pEGFP-C1-NPMmA or pEGFP-C1-NPMwt in presence and absence of LMB. In basal conditions, the mutant A is cytoplasmic (top left) but relocates in the nucleoplasm (top right) following incubation with LMB. eGFP-NPMwt localizes in the nucleoli both in the presence and absence of LMB (middle panel, left and right). The bottom panel shows LMB-treated cells transfected with pEGFP-C1-NPMmA (green) and immunostained with an antinucleolin (C23) monoclonal antibody (Texas red)–labeling nucleoli (arrow). The nuclear membrane is blue (TO-PRO-3 staining). After LMB, the eGFP-NPM mutant A was relocated in the nucleoplasm. Nucleolar exclusion was confirmed by an electronic cut of the nucleolar surface (inset in the bottom right figure). (B) The anti-NPM antibody 376 labels both nucleoli and cytoplasm of OCI/AML3 cells in the absence of LMB (left). Incubation with LMB results in nucleoplasmic relocation of the cytoplasmic NPM (right). Images were collected as described27 using a Zeiss Plan Apochromat 100 ×/1.4 NA oil objective. (C) OCI-AML3 and U937 cell lysates were subjected to immunoprecipitation (IP) with either control IgG or rabbit anti-NPMm (Sil-C) and mouse anti-NPM (Cl. 376): Western blots with anti-Crm1 antibodies are shown in the top panels. Blotting with the anti-NPMm (Sil-C) and anti-NPM (Cl. 376) antibody are also shown (middle and bottom panels). WCL indicates OCI-AML3 whole-cell lysate included as control.
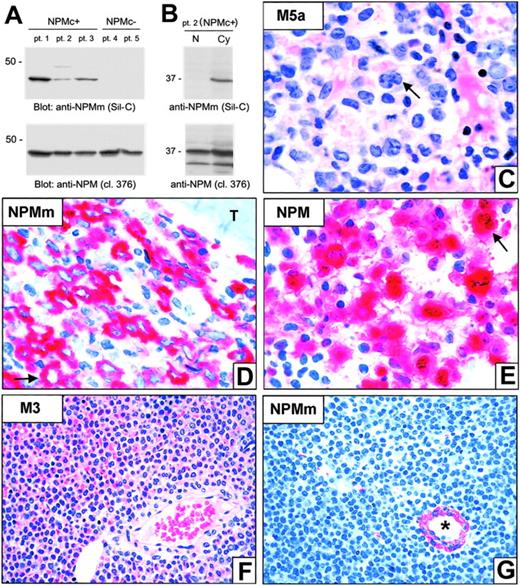
Although the NPM mutant constructs localize exclusively in the cytoplasm in transfected cells,8 subcellular distribution (nuclear vs cytoplasmic) of NPM mutant proteins in primary NPMc+ AML samples could not be investigated because of interference of NPM wild-type protein. To address this issue, we developed a polyclonal antibody (Sil-C) specifically directed against NPM mutants. Upon Western blotting, Sil-C recognized a specific 37-kDa band only in whole-cell lysates from NPMc+ AML primary cells (Figure 5A, top panel; patients 1-3) and not in NPMc– AML samples (patients 4 and 5). Conversely, the anti-NPM monoclonal antibody 376, which also reacts with wild-type NPM, detected the 37-kDa band in all 5 patients (Figure 5A, bottom panel). In patient 2 (carrying NPM mutation A), the Sil-C antibody recognized the 37-kDa band only in the cytoplasm (Figure 5B, top panel) while the 376 antibody detected it in both nuclear and cytoplasmic fractions, as expected (Figure 5B, bottom panel).
These findings were confirmed by immunohistochemical staining of bone marrow biopsies from 10 patients with NPMc+ AML (Figure 5C). Interestingly, the Sil-C antibody not only stained the 7 NPMc+ AMLs carrying the mutation A, but also those carrying the mutation B (1 case) and D (2 cases). In all samples, Sil-C gave cytoplasmic-restricted positivity (Figure 5D), which, in competition experiments, was antagonized by the peptide used as immunogen (not shown). The same peptide did not antagonize cytoplasmic reactivity of a monoclonal antibody (clone 376) directed against a different epitope of NPM (not shown). As expected, immunostaining with monoclonal anti-NPM antibodies recognizing wild-type and mutated NPM17 resulted in nuclear plus cytoplasmic positivity of the leukemic cells (Figure 5E). The Sil-C antibody did not stain leukemic cells in any of the 10 AMLs with an unmutated NPM gene and nucleus-restricted expression of wild-type NPM (NPMc–) used as a negative control (Figure 5F-G).
Export of NPM mutants require 2 NES motifs. Confocal microscope analysis of NIH-3T3 transfected with either pEGFP-C1-NPMmA (left panel), pEGFP-C1-NPM_MUT_A_NO_1st_NES (middle panel), or pEGFP-C1-NPM_MUT_A_NO_2nd_NES (right panel). NPM mutant A containing both NES motifs localizes in the cytoplasm. Disruption of either N-terminal (middle panel) or C-terminal (right panel) NES motif in the NPM mutant A impedes nuclear export of the protein that appears localized in the nucleoplasm (middle and right panels, bottom). Images were collected as described27 using a Zeiss Plan Apochromat 100 ×/1.4 NA oil objective.
Export of NPM mutants require 2 NES motifs. Confocal microscope analysis of NIH-3T3 transfected with either pEGFP-C1-NPMmA (left panel), pEGFP-C1-NPM_MUT_A_NO_1st_NES (middle panel), or pEGFP-C1-NPM_MUT_A_NO_2nd_NES (right panel). NPM mutant A containing both NES motifs localizes in the cytoplasm. Disruption of either N-terminal (middle panel) or C-terminal (right panel) NES motif in the NPM mutant A impedes nuclear export of the protein that appears localized in the nucleoplasm (middle and right panels, bottom). Images were collected as described27 using a Zeiss Plan Apochromat 100 ×/1.4 NA oil objective.
These findings confirm that the endogenous mutated NPM proteins localize exclusively in the cytoplasm of primary NPMc+ AML cells, suggesting that export of mutated NPM protein from the nucleus is highly efficient and more rapid than nuclear import.
NES and tryptophan(s) mutations act in concert to export NPM mutants. Confocal microscope analysis of NIH-3T3 transfected with either pEGFP-C1-NPMwt or different pEGFP-C1-NPM mutants. NPM mutant A localizes exclusively in the cytoplasm. The NPM mutant E, retaining tryptophan 288 but lacking tryptophan 290, partially relocalizes in the nucleoli when the nuclear export of the protein is blocked by LMB treatment. Reinsertion of the tryptophan 290 (NPMmutA, A290W) produced an almost identical effect. When both tryptophans (288 and 290) were reinserted in the mutant A, the eGFP-tagged protein totally localized into the nucleoli, despite the 2 NES motifs, whether LMB was present or not. Images were collected as described27 using a Zeiss Plan Apochromat 100 ×/1.4 NA oil objective.
NES and tryptophan(s) mutations act in concert to export NPM mutants. Confocal microscope analysis of NIH-3T3 transfected with either pEGFP-C1-NPMwt or different pEGFP-C1-NPM mutants. NPM mutant A localizes exclusively in the cytoplasm. The NPM mutant E, retaining tryptophan 288 but lacking tryptophan 290, partially relocalizes in the nucleoli when the nuclear export of the protein is blocked by LMB treatment. Reinsertion of the tryptophan 290 (NPMmutA, A290W) produced an almost identical effect. When both tryptophans (288 and 290) were reinserted in the mutant A, the eGFP-tagged protein totally localized into the nucleoli, despite the 2 NES motifs, whether LMB was present or not. Images were collected as described27 using a Zeiss Plan Apochromat 100 ×/1.4 NA oil objective.
NPM mutants dislocate NPM–wild type in the nucleoplasm and cytoplasm
As all mutated NPM proteins conserve the N-terminal dimerization domain, they may be expected to form heterodimers with wild-type NPM, as occur with wild-type NPM and NPM-containing fusion proteins like NPM-ALK and NPM-MLF1.18,42,44
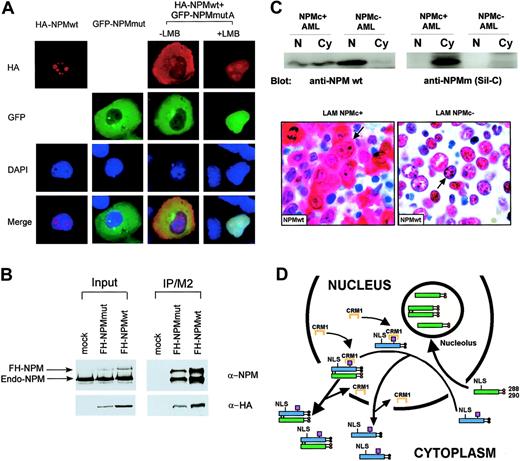
To assess whether delocalized NPM mutant proteins recruit wild-type NPM from nucleoli, we cotransfected H1299 cells with vectors encoding for wild-type NPM-HA and mutant A fused to eGFP. In basal conditions, both mutant (eGFP) and wild-type (anti-HA) NPM were detected in the cytoplasm, indicating NPM mutant A delocalized wild-type protein (Figure 6A). Upon treatment with LMB, both mutant and wild-type NPM shuttled to the nucleoplasm, with some wild-type protein being observed in the nucleoli (Figure 6A). These findings were confirmed by anti-Flag immunoprecipitation of Flag-HA-NPM–mutant A or Flag-HA-NPM-wt–transfected cells, followed by anti-HA and anti-NPM blotting (Figure 6B). These findings demonstrate that, in transfected cells, NPM mutants recruit wild-type protein from the nucleoli and partially delocalize it into the nucleoplasm and cytoplasm.
To investigate whether the wild-type NPM is cytoplasmic-dislocated even in primary AML cells carrying NPM mutations, we used the anti-NPM monoclonal antibody (clone FC-61991) that recognizes the wild-type but not mutant NPM proteins.45 Western blot and immunohistochemistry studies clearly demonstrated the endogenous wild-type NPM was localized both in the nucleus and cytoplasm in NPMc+ AML primary samples while, as expected, was restricted to the nucleus in NPMc– AML cells (Figure 6C).
A putative mechanism of altered nucleo-cytoplasmic traffic of the mutated and wild-type NPM proteins in NPMc+ AML is depicted in Figure 6D.
Discussion
The present study shows how specific mutations on the NPM exon-12 sequences severely impair NPM nucleo-cytoplasmic trafficking in transfected cells, AML cell line OCI/AML3 (carrying exon-12 mutation A), and primary NPMc+ AML cells. Consequent to the mutations, 2 major alterations occur at the C-terminus of NPM leukemic mutants: (1) generation of a leucine-rich NES motif that reinforces the Crm1-dependent nuclear export of mutated NPM proteins; and (2) loss of the C-terminus tryptophan(s) residues that are normally required for NPM binding to the nucleoli.
NPM mutants localize exclusively in the cytoplasm of leukemic cells. (A-B) Western blot analysis (Sil-C vs 376 mAb) of whole-cell lysates from either NPMc+ or NPMc– AML cells (A) and of nuclear (N) and cytoplasmic (Cy) cell fractions from a patient with AML bearing NPM mutation A (B). Sil-C recognizes specifically the mutated NPM protein in the cytoplasmic fraction of NPMc+ primary leukemic cells. Results are representative of at least 3 independent experiments. (C-E) Bone marrow biopsy from a NPMc+ AML bearing mutation A. (C) Typical appearance of FAB-M5a type AML; the arrow points to a large leukemic cell with kidney-shaped nucleus. (D) The Sil-C antibody that recognizes specifically the NPM mutant (NPMm) shows a cytoplasmic-restricted positivity of leukemic cells (arrow); T indicates a bone trabecula. (E) Monoclonal anti–NPM 376 that recognizes both the wild-type and mutant NPM proteins labels leukemic cells both in the cytoplasm and the nucleus (arrow). (F-G) Bone marrow biopsy from a case of promyelocytic leukemia lacking NPM mutations (NPMc–). (F) Typical appearance of AML FAB-M3 (hematoxylineosin). (G) The Sil-C antibody does not stain leukemic cells (devoid of mutated NPM protein) and only cross-reacts with a vessel wall; the asterisk indicates the lumen of the vessel (APAAP technique; hematoxylin counterstain). Images were collected using an Olympus BX51 microscope and a UPlanFl 100 ×/1.3 NA oil (C-E) or a UPlanApo 40 ×/0.85 (F-G) objective.
NPM mutants localize exclusively in the cytoplasm of leukemic cells. (A-B) Western blot analysis (Sil-C vs 376 mAb) of whole-cell lysates from either NPMc+ or NPMc– AML cells (A) and of nuclear (N) and cytoplasmic (Cy) cell fractions from a patient with AML bearing NPM mutation A (B). Sil-C recognizes specifically the mutated NPM protein in the cytoplasmic fraction of NPMc+ primary leukemic cells. Results are representative of at least 3 independent experiments. (C-E) Bone marrow biopsy from a NPMc+ AML bearing mutation A. (C) Typical appearance of FAB-M5a type AML; the arrow points to a large leukemic cell with kidney-shaped nucleus. (D) The Sil-C antibody that recognizes specifically the NPM mutant (NPMm) shows a cytoplasmic-restricted positivity of leukemic cells (arrow); T indicates a bone trabecula. (E) Monoclonal anti–NPM 376 that recognizes both the wild-type and mutant NPM proteins labels leukemic cells both in the cytoplasm and the nucleus (arrow). (F-G) Bone marrow biopsy from a case of promyelocytic leukemia lacking NPM mutations (NPMc–). (F) Typical appearance of AML FAB-M3 (hematoxylineosin). (G) The Sil-C antibody does not stain leukemic cells (devoid of mutated NPM protein) and only cross-reacts with a vessel wall; the asterisk indicates the lumen of the vessel (APAAP technique; hematoxylin counterstain). Images were collected using an Olympus BX51 microscope and a UPlanFl 100 ×/1.3 NA oil (C-E) or a UPlanApo 40 ×/0.85 (F-G) objective.
NPM mutant A acts as a dominant-negative on wild-type NPM. (A) Cotransfection of H1299 cells with pEGFP-C1-NPMmA and HA-NPM wild-type. About 30% of cells were transfected and in about 70% of transfected cells the NPM mutant causes partial recruitment of the wild-type NPM from nucleoli to the nucleoplasm and the cytoplasm. (B) Both FH-NPMwt and FH-NPM mutant A interact with the endogenous NPM in H1299 cells. FH-NPM wt and FH-NPM mutant A plasmids were stably transfected into H1299 cells. In the left panel, 5% of the whole-cell lysates from stably transfected FH-NPMwt, FH-NPM mutant A, or control H1299 cells were subjected to Western blot with either α-NPM or α-HA. In the right panel, the rest 95% of whole cell lysates were immunoprecipitated with α-flag (M2), followed by Western blot with either α-NPM or α-HA. (C) Endogenous wild-type NPM protein is dislocated in the cytoplasm of NPMc+ AML primary samples. In the top panel, nuclear and cytoplasmic extracts from either NPMc+ or NPMc– AML primary patient cells were subjected to Western blot analysis with either the anti-NPM mouse monoclonal antibody (clone FC-61991) from Invitrogen (anti-NPM wt) or the Sil-C antibody (anti-NPMm). A strong signal for NPMwt was evident in both the nuclear and cytoplasmic fraction of AML cells carrying NPM mutation but only in the nuclear fraction of NPMc–AML cells. The lower panel shows immunohistochemical study of 2 different AML samples with the anti-NPMwt antibody. The NPMc+ AML sample expresses the wild-type NPM both in the nucleus and cytoplasm (arrow). In contrast, the NPMc–AML sample shows the expected nucleus-restricted expression of wild-type NPM (arrow). Images were collected using an Olympus BX61 microscope and a UPlanFl 100 ×/1.3 NA objective. (D) Hypothetical mechanism of altered nucleo-cytoplasmic traffic of NPM mutant A and wild-type NPM. Red circles indicate tryptophan residues; black circles, mutated tryptophans; magenta boxes, NES motif; blue boxes, mutated NPM protein; and green boxes, wild-type NPM protein. To simplify, the scheme shows only the binding of Crm1 to the mutant NES but not to the physiologic one, nor to the full ternary complex (Crm1-NES-ranGTP). NLS indicates nuclear localization signal.
NPM mutant A acts as a dominant-negative on wild-type NPM. (A) Cotransfection of H1299 cells with pEGFP-C1-NPMmA and HA-NPM wild-type. About 30% of cells were transfected and in about 70% of transfected cells the NPM mutant causes partial recruitment of the wild-type NPM from nucleoli to the nucleoplasm and the cytoplasm. (B) Both FH-NPMwt and FH-NPM mutant A interact with the endogenous NPM in H1299 cells. FH-NPM wt and FH-NPM mutant A plasmids were stably transfected into H1299 cells. In the left panel, 5% of the whole-cell lysates from stably transfected FH-NPMwt, FH-NPM mutant A, or control H1299 cells were subjected to Western blot with either α-NPM or α-HA. In the right panel, the rest 95% of whole cell lysates were immunoprecipitated with α-flag (M2), followed by Western blot with either α-NPM or α-HA. (C) Endogenous wild-type NPM protein is dislocated in the cytoplasm of NPMc+ AML primary samples. In the top panel, nuclear and cytoplasmic extracts from either NPMc+ or NPMc– AML primary patient cells were subjected to Western blot analysis with either the anti-NPM mouse monoclonal antibody (clone FC-61991) from Invitrogen (anti-NPM wt) or the Sil-C antibody (anti-NPMm). A strong signal for NPMwt was evident in both the nuclear and cytoplasmic fraction of AML cells carrying NPM mutation but only in the nuclear fraction of NPMc–AML cells. The lower panel shows immunohistochemical study of 2 different AML samples with the anti-NPMwt antibody. The NPMc+ AML sample expresses the wild-type NPM both in the nucleus and cytoplasm (arrow). In contrast, the NPMc–AML sample shows the expected nucleus-restricted expression of wild-type NPM (arrow). Images were collected using an Olympus BX61 microscope and a UPlanFl 100 ×/1.3 NA objective. (D) Hypothetical mechanism of altered nucleo-cytoplasmic traffic of NPM mutant A and wild-type NPM. Red circles indicate tryptophan residues; black circles, mutated tryptophans; magenta boxes, NES motif; blue boxes, mutated NPM protein; and green boxes, wild-type NPM protein. To simplify, the scheme shows only the binding of Crm1 to the mutant NES but not to the physiologic one, nor to the full ternary complex (Crm1-NES-ranGTP). NLS indicates nuclear localization signal.
The NPM wild-type protein contains a putative hydrophobic leucine-rich NES motif (I-xx-P-xx-L-x-L) within residues 94-102 which is conserved from Xenopus laevis to humans.40 Despite the presence of this motif, the NPM wild-type protein is mainly localized in the nucleoli suggesting that, in physiologic conditions, protein import greatly predominates over nuclear export. Interestingly, rat41 and human40 NPM wild-type protein mutants lacking the 2 tryptophans 288 and 290 delocalize in the nucleoplasm but not in the cytoplasm,40,41 strongly suggesting that the additional NES motif reinforces the leukemic NPM mutant ability to be exported from the nucleus to the cytoplasm. As the NES motif at the C-terminus of leukemic NPM mutant A is absent in the NPM wild-type protein, enhanced export could be due to the additive effects of the 2 NES and/or to an increased affinity of the new NES motif for Crm1. The results presented in this paper indicates that cytoplasmic accumulation of NPM mutants depends on the additive effect of the newly generated C-terminal NES motif.
The tryptophans 288 and 290 both appear to be important for nucleolar localization, with tryptophan 290 being perhaps more critical since it is mutated in all NPM mutants so far identified. Maximal inhibition of nucleolar binding and nucleoplasmic delocalization of mutants is observed when both tryptophans are mutated. Notably, mutations affecting both tryptophans 288 and 290 were detected in 20 (69%) of 29 NPM mutants identified to date, and account for about 95% of all tryptophan mutations found in NPM leukemic mutants.
The correlation between the type of NES motif and mutations of tryptophans 288 and 290 is intriguing. The observation that the NES motif most commonly found in the NPM mutants (ie, L-xxx-V-xx-V-x-L) is always associated with mutations of both tryptophans and that tryptophan 288 is retained only in the NPM mutants carrying variants of the most common NES motif (ie, substitution of the valine at the second position of NES with leucine, phenylalanine, cysteine or methionine), suggest a functional difference between the NES motifs.
Taken together, our data clearly show that, for cytoplasmic accumulation of NPM to occur, the 2 NES motifs and tryptophan(s) mutations must act in concert. This view clearly differs from that of Nagakawa et al,26 who claimed the NES motif at the C-terminus was the only player in cytoplasmic dislocation of NPM leukemic mutants. The mechanism of altered nucleo-cytoplasmic traffic in NPMc+ AML cells also emerges as different to what den Besten et al45 recently observed in transfected cells. In MT-Arf cells, leukemic NPM mutant A relocation after incubation with LMB is reported to be nucleolar, while we clearly demonstrate relocation of NPM mutant A by Crm1-inhibitors is nucleoplasmic in transfected NIH-3T3 cells. Several points need to be borne in mind when analyzing these discrepancies. Like Nakagawa et al,26 den Besten et al45 appear to disregard or underestimate the role of mutated tryptophan(s) 288 and 290 in the altered nucleo-cytoplasmic transport of NPM leukemic mutants. Artificial interference by the Arf protein in MT-Arf cells used by den Besten et al45 may be another factor. Finally, den Besten et al45 show that much wild-type NPM protein is delocalized in the cytoplasm by the NPM mutant A. Our immunohistologic studies of bone marrow biopsies using anti-NPM antibodies, including reagents specific for the wild-type and mutated NPM proteins, confirm the presence of cytoplasmic-dislocated wild-type NPM in primary NPMc+ AML cells, but also reveal that a residue of wild-type NPM remains in the nucleus (Figure 6C, bottom panels). Further studies are required to exactly quantify by fluorescence-based techniques the ratio of wild-type to mutated NPM in the subcellular compartments of NPMc+ primary leukemic cells.
Based on these findings, the following scenario can be envisaged. Mutated NPM proteins that retain the 2 nuclear localization signals (NLSs) enter the nucleus. Their capability to bind nucleoli is hindered completely (when both tryptophans are mutated) or partially (when only tryptophan 290 is altered). Nucleoplasmic NPM mutants may become easily available for binding to Crm1 roaming in the nucleoplasm46 through the NES motifs. NPM mutants accumulate in the cytoplasm most likely because the NES-Crm1–mediated nuclear export of mutants is more efficient than nuclear import. The NPM leukemic mutants cause in turn partial delocalization of the NPM wild-type protein from the nucleus to the cytoplasm.
Our demonstration that antibodies can be produced to react with mutant, but not with wild-type, NPM proteins is of biological interest and potential clinical relevance. It might be feasible in the future to use intracellular antibodies (“intrabodies”)47 against NPM mutants to ablate protein function as an alternative to gene-based knockout technologies which do not appear to discriminate between wild-type and mutated NPM encoding RNAs (B.F. and A.L., unpublished observations, June 2005). The OCI/AML3 cell line may well serve as a preclinical model for screening intrabodies and other compounds designed to interfere with the NPM-altered nucleo-cytoplasmic traffic.
The NPM wild-type protein plays a critical role in the control of hematopoiesis (especially erythropoiesis) during embryonic development,15 and is involved in regulation of the Arf-p53 pathway9,11,14 and centrosome duplicaton.10,40 Our finding that in transfected cells NPM mutants recruit wild-type NPM to the nucleoplasm and cytoplasm suggests they interfere with native protein functions and could contribute to AML. However, exactly how the mutants exert their putative leukemogenic action remains elusive. Den Besten et al45 showed that NPM mutant proteins dislocate both wild-type NPM and the Arf protein into the cytoplasm, but were unable to demonstrate any transforming activity of the NPM mutants in NIH-3T3 and in Atm-null MEF cells. These findings suggest that NPM mutants could exert their transforming activity in a tissue-specific manner, an issue that needs to be addressed using appropriate animal models. Animal models show that AML may arise as result of the cooperative effect of different genetic lesions (eg, FLT3-ITD mutations in combination with molecular alterations, such PML-RAR alpha or AML1-ETO fusion genes).1 We previously demonstrated that FLT3-ITD mutations in AML with normal karyotype occur twice as frequently in cases with NPM mutations as in those without.8 Consequently, a major focus of future studies should be to determine whether interactions between FLT3-ITD and NPM mutations contribute to the leukemogenesis. Although at present there is no evidence for the leukemogenic potential of NPM mutations, if it should be demonstrated in the future, then retargeting NPM mutants to their physiologic cell compartment (nucleoli) might be a potential therapeutic strategy.36,37 This paper clearly shows that, to be successful, retargeting has to overcome 2 obstacles, the new NES motif and the mutated tryptophan(s) at the mutant C-terminus.
Prepublished online as Blood First Edition Paper, February 2, 2006; DOI 10.1182/blood-2005-11-4745.
Supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC). L.P. is a Special Fellow of the Leukemia and Lymphoma Society. N.B. is supported by the Federazione Italiana per la Ricerca sul Cancro (FIRC).
Two of the authors (B.F. and C.M.) have applied for a patent, related to the work described in the present study, on the clinical use of NPM mutants.
N.B. and J.S. contributed equally to this work.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are indebted to Roberta Pacini, Alessia Tabarrini, and Barbara Verducci for their skillfull technical assistance, and to Claudia Tibidò for her excellent secretarial assistance. We would also like to thank Dr Geraldine Anne Boyd for her help in editing this paper.

![Figure 1. Nuclear export of NPM mutants is NES dependent. (A-B) H1299 cells transfected with cDNA encoding for the NPMwt and the NPM mutant proteins A to D8 in the absence (A) and presence (B) of LMB. eGFP-NPMwt localizes in the nucleoli both in the presence and absence of LMB. In the absence of LMB, all GFP mutants (A-D) show the expected aberrant cytoplasmic location (A). The same eGFP-mutants are completely relocated in the nucleus in the presence of LMB (B). (C-E) Time-course analysis of LMB induced nuclear retention of eGFP-NPMmutA in NIH-3T3 cells. (C) Confocal microscope images of NIH-3T3 cells at the indicated time points. ○ represents the regions (regions of interest [ROIs]) where the mean fluorescence emission of eGFP-NPMmutA was calculated in the experiment (eg, nucleus [N], cytoplasm [Cy], and the Golgi apparatus [G]). (D) Time-course measurements of mean fluorescence in the selected areas (ROIs) of NIH-3T3 cells. LMB addition produced a loss of fluorescence in the cytoplasm and Golgi apparatus and a concomitant increase in the mean fluorescence of the nucleoplasm. ImageJ software was used for the generation of ROIs and time-course analysis. (E) Western blot analysis of eGFP-NPMmutA subcellular distribution in NIH-3T3 cells with anti-GFP monoclonal antibody. LMB treatment induces a time-dependent accumulation of eGFP-NPMmutA in the pellet fractions (P). Purity of the subcellular fractions was assessed by stripping the membrane and reblotting with an anti–β-tubulin antibody (bottom panel). Only a nonsignificant contamination was detected for the overexpressed proteins (as evident in the GFP blotting) (middle panel). In untreated cells, eGFP-NPMmutA (except for the contaminant part) was found only in the cytoplasmic fractions (Cy). The experiment was conducted in absence of cycloheximide so that continuous presence of GFP-NPMmA in the cytoplasmic fraction during LMB treatment was detected with time.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/11/10.1182_blood-2005-11-4745/4/m_zh80110696380001.jpeg?Expires=1767710637&Signature=3hXC8sncdh5GyhJCWfvu-v3iWiKyWMPBF8tRFGvjYzZv5BqUnLHLzqVqUXl5kMYwHh1oKLrqEQZsBPWfzFj2PsnXR8z62Y3rwUp2tUFZPBPZnqxDkHmjbIYmtLuVNyf5ac0XN~aqvkpoXHCrepOyCOqXRVPr6l-Wg87piU38dxqD2IHvXm1qEwXEj2hAm~ndOjO-rXaoj0Tm0ZkZ2uAcrpUUln-FvI1a8rMfD9-z3Xy2hJJc5V1rH0Qui4e8UpyE8zHLi1XXzE2nUiQXeML9DoTS7veW-6bnGuJb~dTmiP9so7gQHQ8fRl4inyq2XOHdbMyZRtYNkrKpBAyv5mHi8w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal