To the editor:
The JAK2V617F mutation is found in most patients with polycythemia vera (PV) and 50% to 60% of those with essential thrombocythemia (ET). JAK2V617F-homozygous precursors arise through mitotic recombination, form larger clones in PV compared with ET,1 and may play a causal role in PV phenotypes.2,3 However, acquisition of homozygosity is not sufficient to cause PV, because many ET patients also harbor homozygous-mutant clones1 and several studies have suggested that JAK2V617F, especially when homozygous, may not confer an advantage to hematopoietic stem cells (HSCs).3-5 Moreover, many PV and ET patients have multiple independently acquired homozygous-mutant clones, with most remaining small.1 PV is distinguished from ET by expansion of 1 dominant JAK2V617F-homozygous subclone. Here we investigated whether subclone expansion reflects acquisition of additional genetic lesions conferring a clonal advantage.
We studied 2 patients with chronic-phase PV (see supplemental Table 1 on the Blood Web site) and large JAK2V617F-homozygous clones.1 Genotyping of burst-forming unit-erythroid (BFU-E) colonies for JAK2V167F and microsatellite markers (as previously described1 ) demonstrated that both patients had 3 detectable JAK2V617F-homozygous subclones, 1 of which was 9 to 15 times larger than minor subclones in the same patient (Figure 1A). Serial assays confirmed that these subclones persisted for approximately 2 years and that the dominance of 1 subclone remains stable (Figure 1B). To investigate whether this dominance arose in HSCs or later progenitors, we isolated highly purified hematopoietic progenitors from patient PV1 (supplemental Methods). Colony genotyping demonstrated that the major JAK2V617F-homozygous subclone in BFU-Es also predominated in HSCs, common myeloid progenitors, granulocyte-monocyte progenitors, and megakaryocyte-erythroid progenitors (Figure 1C), suggesting that this dominance arose early in hematopoiesis.
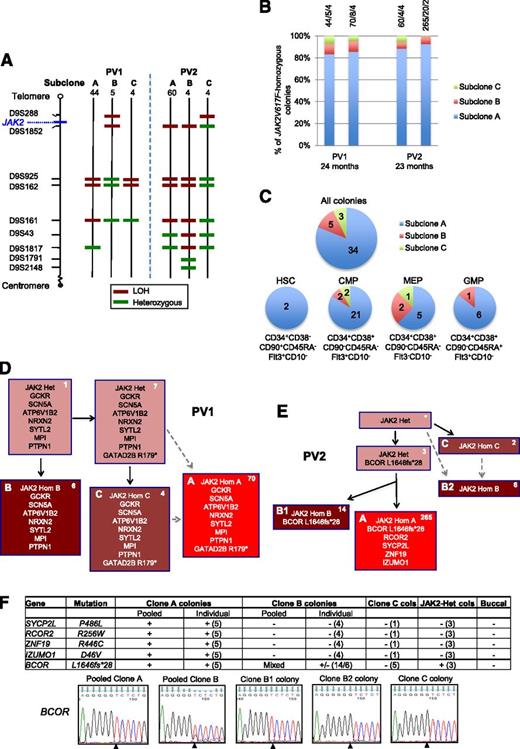
Investigation of mechanisms for expansion of JAK2V617F-homozygous subclones in patients PV1 and PV2. (A) Microsatellite mapping of breakpoints for loss of heterozygosity (LOH) on 9p in JAK2V617F-homozygous BFU-E colonies from patients PV1 and PV2 (reported previously1 ). For microsatellite markers on chromosome 9 (left), 3 patterns were observed in colonies for each patient, indicating 3 distinct LOH breakpoints. Numbers of colonies per subclone are shown at the top. Green denotes a heterozygous marker; red denotes LOH. (B) Persistence of multiple JAK2V617F-homozygous subclones over time. JAK2V617F-homozygous BFU-E colonies grown at 2 time points (A and B) at 0.01 U/mL erythropoietin were genotyped for microsatellite markers on 9p. Bars are divided according to the number of colonies corresponding to subclones A, B, and C (panel A), with absolute numbers at the top (A/B/C). The time between assays is shown at the bottom. (C) Presence of multiple JAK2V617F-homozygous subclones in hematopoietic progenitor compartments. Patient PV1 was used because a high JAK2V617F allele burden (99% in granulocytes) permitted growth of many JAK2V617F-homozygous colonies. Colonies were cultured from fluorescence-activated cell sorted peripheral blood progenitors, and JAK2V617F-homozygous colonies were genotyped for microsatellite markers on 9p. Each pie chart is divided according to the number of colonies corresponding to subclones A, B, and C from panel A; for all colonies combined (top), and for HSC, common myeloid progenitor (CMP), megakaryocyte-erythroid progenitor (MEP), or granulocyte-monocyte progenitor (GMP) populations separately (bottom). Phenotypic definitions are shown. (D) Colony hierarchy for PV1. Mutant genes are shown in each box with the corresponding number of colonies at the top right in white. Pink boxes denote JAK2V617F-heterozygous colonies; red denotes JAK2V617F-homozygous, with different subclones shown by letters and red shades. Subclones C and A may have occurred sequentially (C then A, according to the LOH breakpoints) or may have arisen from the same JAK2V617F-heterozygous precursor (gray dashed arrows). Subclone B lacks the GATAD2B mutation and must have acquired 9p LOH independently. Full mutation details are shown in supplemental Table 2. (E) Colony hierarchy for PV2. Numbers of colonies from each subclone at a single time point are shown at the top right of each box in white, and the additional mutations detected in validation studies (on pooled or individual colonies, panel F) are shown. The BCOR wild-type clone B colonies are likely to represent an independent subclone (“B2”) with a similar 9p LOH breakpoint to the BCOR-mutant subclone (“B1”) (A,E: resolution of microsatellite mapping is 2.5 Mb). Subclones C and B2 may have arisen independently but could also have arisen sequentially (C then B2, according to the LOH breakpoints; gray dashed arrows). Subclone B1 has a more distal breakpoint than subclone A and cannot be an intermediate stage in its development. However, subclones A and B1 could have diverged either before 9p LOH or after an initial proximal LOH event. Additional variants were called by exome sequencing in both subclones A and B but were not pursued for validation (supplemental Table 2). *No colonies with this genotype were identified, but the presence of this intermediate can be inferred. Het, heterozygous; Hom, homozygous. (F) Genotyping data of 5 validated mutations in PV2 in pooled colonies from JAK2V617F-homozygous subclones, in individual colonies (number tested appears in parentheses) and in buccal DNA, from which the hierarchy in panel (E) is derived. +, presence of mutation; −, absence of mutation; MT, mutant allele; WT, wild-type allele. For clone C, 1 colony was genotyped for all mutations and an additional 4 from an earlier time point were genotyped for BCOR; given that all of these colonies are BCOR wild-type, the clone must be independent and wild-type for the other mutations. Capillary sequencing traces for the BCOR mutation are shown at the bottom; the mutation (del g) is present in pooled clone A and individual clone B1 colonies; pooled clone B colonies show a mixed trace; the wild-type sequence is present in clone B2 and C colonies. Note that the mutation is hemizygous (BCOR is X-linked; the patient is male).
Investigation of mechanisms for expansion of JAK2V617F-homozygous subclones in patients PV1 and PV2. (A) Microsatellite mapping of breakpoints for loss of heterozygosity (LOH) on 9p in JAK2V617F-homozygous BFU-E colonies from patients PV1 and PV2 (reported previously1 ). For microsatellite markers on chromosome 9 (left), 3 patterns were observed in colonies for each patient, indicating 3 distinct LOH breakpoints. Numbers of colonies per subclone are shown at the top. Green denotes a heterozygous marker; red denotes LOH. (B) Persistence of multiple JAK2V617F-homozygous subclones over time. JAK2V617F-homozygous BFU-E colonies grown at 2 time points (A and B) at 0.01 U/mL erythropoietin were genotyped for microsatellite markers on 9p. Bars are divided according to the number of colonies corresponding to subclones A, B, and C (panel A), with absolute numbers at the top (A/B/C). The time between assays is shown at the bottom. (C) Presence of multiple JAK2V617F-homozygous subclones in hematopoietic progenitor compartments. Patient PV1 was used because a high JAK2V617F allele burden (99% in granulocytes) permitted growth of many JAK2V617F-homozygous colonies. Colonies were cultured from fluorescence-activated cell sorted peripheral blood progenitors, and JAK2V617F-homozygous colonies were genotyped for microsatellite markers on 9p. Each pie chart is divided according to the number of colonies corresponding to subclones A, B, and C from panel A; for all colonies combined (top), and for HSC, common myeloid progenitor (CMP), megakaryocyte-erythroid progenitor (MEP), or granulocyte-monocyte progenitor (GMP) populations separately (bottom). Phenotypic definitions are shown. (D) Colony hierarchy for PV1. Mutant genes are shown in each box with the corresponding number of colonies at the top right in white. Pink boxes denote JAK2V617F-heterozygous colonies; red denotes JAK2V617F-homozygous, with different subclones shown by letters and red shades. Subclones C and A may have occurred sequentially (C then A, according to the LOH breakpoints) or may have arisen from the same JAK2V617F-heterozygous precursor (gray dashed arrows). Subclone B lacks the GATAD2B mutation and must have acquired 9p LOH independently. Full mutation details are shown in supplemental Table 2. (E) Colony hierarchy for PV2. Numbers of colonies from each subclone at a single time point are shown at the top right of each box in white, and the additional mutations detected in validation studies (on pooled or individual colonies, panel F) are shown. The BCOR wild-type clone B colonies are likely to represent an independent subclone (“B2”) with a similar 9p LOH breakpoint to the BCOR-mutant subclone (“B1”) (A,E: resolution of microsatellite mapping is 2.5 Mb). Subclones C and B2 may have arisen independently but could also have arisen sequentially (C then B2, according to the LOH breakpoints; gray dashed arrows). Subclone B1 has a more distal breakpoint than subclone A and cannot be an intermediate stage in its development. However, subclones A and B1 could have diverged either before 9p LOH or after an initial proximal LOH event. Additional variants were called by exome sequencing in both subclones A and B but were not pursued for validation (supplemental Table 2). *No colonies with this genotype were identified, but the presence of this intermediate can be inferred. Het, heterozygous; Hom, homozygous. (F) Genotyping data of 5 validated mutations in PV2 in pooled colonies from JAK2V617F-homozygous subclones, in individual colonies (number tested appears in parentheses) and in buccal DNA, from which the hierarchy in panel (E) is derived. +, presence of mutation; −, absence of mutation; MT, mutant allele; WT, wild-type allele. For clone C, 1 colony was genotyped for all mutations and an additional 4 from an earlier time point were genotyped for BCOR; given that all of these colonies are BCOR wild-type, the clone must be independent and wild-type for the other mutations. Capillary sequencing traces for the BCOR mutation are shown at the bottom; the mutation (del g) is present in pooled clone A and individual clone B1 colonies; pooled clone B colonies show a mixed trace; the wild-type sequence is present in clone B2 and C colonies. Note that the mutation is hemizygous (BCOR is X-linked; the patient is male).
Exome sequencing (supplemental Methods) was next performed to search for mutations present in major, but not minor, JAK2V617F-homozygous subclones. High tumor-burden samples were used to maximize mutation detection. Variant validation and colony genotyping were performed by capillary sequencing. For PV1, granulocyte DNA (JAK2V617F allele burden 99%) was used for exome sequencing and validation of somatic mutations in 8 genes plus JAK2V617F (Figure 1D; supplemental Table 2). Of these, the exact nonsense variant in transcriptional repressor GATAD2B has been identified in 3 patients with acute myeloid leukemia, whereas none of the other genes is recurrently mutated in myeloid malignancies.6 The GATAD2B mutation was detected in the major JAK2V617F-homozygous subclone but also in 1 minor JAK2V617F-homozygous subclone (C), together with a subset of JAK2V617F-heterozygous colonies (Figure 1D).
For PV2, granulocyte DNA had a low tumor burden (JAK2V617F allele, 18.1%), reducing the sensitivity of exome sequencing. To circumvent this issue, we pooled BFU-E colonies from subclones A and B and performed independent exome sequencing on these subclones. Variants identified in both (supplemental Table 3) were not pursued. Of the variants in subclone A alone, 5 mutations (plus JAK2V617F) were validated by capillary sequencing (Figure 1E-F; supplemental Table 2). Of these genes, BCOR shows recurrent frameshift mutations in acute myeloid leukemia and myelodysplasias,7 whereas the others are not recurrently mutated in hematological malignancies.6 The hemizygous BCOR mutation was detected in the dominant subclone A but also in a subset of colonies from minor subclone B (Figure 1E-F).
In summary, the dominant JAK2V617F-homozygous subclone in both PV patients harbored a mutation in an additional gene that is mutated in myeloid malignancies. Interestingly, both GATAD2B and BCOR are transcriptional repressors associated with the Mi-2/NuRD complex,8,9 raising the possibility that aberrant histone deacetylation is advantageous to JAK2V617F-homozygous cells. However, both mutations were also present in a minor JAK2V617F-homozygous subclone and cannot account for the dominance of the larger clones.
Our results therefore indicate the absence of known “driver” mutations specific for the dominant JAK2V617F-homozygous subclone. Expansion of the latter may nonetheless reflect genetic differences between dominant and minor subclones. For example, we cannot exclude additional mutations in regions poorly covered by exome sequencing, epigenetic differences, or disadvantageous mutations in the minor subclones. Alternatively, other mutations restricted to the dominant subclones may represent rarely mutated cancer genes, or extension of 9p LOH could provide an advantage in certain cases (eg, PV1). However our data raise the alternative possibility that in at least some patients, nongenetic stochastic mechanisms may favor individual subclones by chance and establish long-term subclone dominance. In this scenario, subclone expansion would not require a genetic advantage but instead may reflect the unique combination of environmental inputs experienced by a particular stem cell.10
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank the core facility of the Cancer Genome Project, Wellcome Trust Sanger Institute, Hinxton, Cambridge, and the flow cytometry facility of the Cancer Research UK Cambridge Institute for technical assistance. Samples were provided by the Cambridge Blood and Stem Cell Biobank, which is supported by the Cambridge National Institute for Health Research Biomedical Research Centre and the Cambridge Experimental Cancer Medicine Centre, Cambridge, United Kingdom. The work in A.R.G.’s laboratory is supported by Leukemia and Lymphoma Research, Cancer Research UK, the Wellcome Trust, the Medical Research Council, the Kay Kendall Leukaemia Fund, the Cambridge National Institute for Health Research Biomedical Research Center, the Cambridge Experimental Cancer Medicine Centre, and the Leukemia and Lymphoma Society of America. This work was supported by the Kay Kendall Leukaemia Fund (A.L.G., J.N.); and a postdoctoral fellowship from the Canadian Institutes of Health Research and a Lady Tata Memorial Trust International Award for Research in Leukaemia (D.G.K.). P.J.C. is a Wellcome Trust senior clinical fellow.
Contribution: A.L.G. performed the research and wrote the paper; J.N., C.E.M., E.P., and P.J.C. performed exome sequencing and data analysis; E.J.B. prepared samples and performed quantitative polymerase chain reactions and quality control; D.G.K. performed fluorescence-activated cell sorting experiments; A.R.G. directed the research and wrote the paper; and all authors had the opportunity to review the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: A. R. Green, Cambridge Institute for Medical Research, Hills Rd, Cambridge, United Kingdom; e-mail: arg1000@cam.ac.uk.