Key Points
CXXC5 inhibits Wnt signaling and is a candidate tumor suppressor in AML.
Low CXXC5 expression is an independent prognostic factor in AML.
Abstract
The gene CXXC5 on 5q31 is frequently deleted in acute myeloid leukemia (AML) with del(5q), suggesting that inactivation of CXXC5 might play a role in leukemogenesis. Here, we investigated the functional and prognostic implications of CXXC5 expression in AML. CXXC5 mRNA was downregulated in AML with MLL rearrangements, t(8;21) and GATA2 mutations. As a mechanism of CXXC5 inactivation, we found evidence for epigenetic silencing by promoter methylation. Patients with CXXC5 expression below the median level had a lower relapse rate (45% vs 59%; P = .007) and a better overall survival (OS, 46% vs 28%; P < .001) and event-free survival (EFS, 36% vs 21%; P < .001) at 5 years, independent of cytogenetic risk groups and known molecular risk factors. In gene-expression profiling, lower CXXC5 expression was associated with upregulation of cell-cycling genes and codownregulation of genes implicated in leukemogenesis (WT1, GATA2, MLL, DNMT3B, RUNX1). Functional analyses demonstrated CXXC5 to inhibit leukemic cell proliferation and Wnt signaling and to affect the p53-dependent DNA damage response. In conclusion, our data suggest a tumor suppressor function of CXXC5 in AML. Inactivation of CXXC5 is associated with different leukemic pathways and defines an AML subgroup with better outcome.
Introduction
Outcome of younger adults (<60 years) with acute myeloid leukemia (AML) remains poor, with survival rates of only 40% to 45%,1,2 highlighting the need for new therapeutic strategies. Several prognostically significant recurrent mutations and aberrant signaling pathways have been identified in AML, which allowed for improved risk stratification and development of targeted therapies. Further refinement of relevant molecular alterations in different AML subgroups might eventually result in more individual treatment approaches and potentially improve outcome.
The CXXC-type zinc finger protein CXXC5 has exhibited tumor-suppressor functions in solid tumor cells.3 In addition, CXXC5 has been demonstrated to negatively regulate the canonical Wnt/β-catenin signaling pathway in nontumorous tissues.4,5 Wnt activation has been implicated in leukemic transformation6,7 and was shown to promote proliferation and survival of leukemic cells in vitro.8
The CXXC motif is found in a small number of proteins, most of which are involved in epigenetic modification, like the DNA methyltransferase DNMT1 (CXXC9), histone methyltransferases (MLL [CXXC7], MLL2 [CXXC12]), the methylcytosine dioxygenase TET1 (CXXC6), and histone demethylases (KDM2A [CXXC8], KDM2B [CXXC2]). MLL plays an important role in development and progression of acute leukemias.9 DNMT110-12 and TET112,13 have more recently been related to leukemogenesis in addition to enzyme family members DNMT3A14 and TET2.15 Inactivation of CXXC4, the paralog of CXXC5, has been associated with tumor progression and inferior outcome in renal cell carcinoma.16
CXXC5 is located on 5q31.2, a region recurrently deleted in AML, and was found to be downregulated in AML with del(5q).17 In addition, somatic microdeletions containing CXXC5 (and CXXC4) have been described in patients with AML and myelodysplastic syndrome (MDS),18,19 further supporting involvement of CXXC5 in leukemogenesis. CXXC5 has been demonstrated to be induced by treatment with all-trans retinoic acid in acute promyelocytic leukemia (APL).20 It was further shown that CXXC5 expression increases during cytokine-driven maturation of hematopoietic precursors, indicating a functional role of CXXC5 in normal myelopoiesis.20 A recent study reported heterogeneous CXXC5 expression levels in a small cohort of de novo and secondary AML and suggested an impact of CXXC5 expression on chemosensitivity and outcome.21
Given the functional implications of CXXC5 in normal myeloid and leukemic development, its putative role as a tumor suppressor, and its potential prognostic relevance, CXXC5 is an interesting candidate gene in myeloid malignancies. Therefore, we aimed to study the functional and prognostic impact of CXXC5 in AML.
Patients and methods
Patient samples
We analyzed microarray-based CXXC5 expression in 529 diagnostic bone marrow (BM) mononuclear cell samples from consecutive AML patients enrolled in Dutch-Belgian-Swiss Hemato-Oncology Cooperative Group (HOVON/SAKK) trials22,23 (see supplemental Material, available on the Blood Web site; median age, 46 years [range, 15-77]). Samples contained at least 80% blasts. From 231 cases of this cohort (median age, 48 years [range, 15-77]), CXXC5 mRNA expression was also assessed by reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) and compared with 19 CD34+ BM samples obtained from patients undergoing hip replacement (median age, 64 years). We also analyzed microarray data from 351 normal karyotype (NK)-AML patients derived from the Microarray Innovations in LEukemia (MILE) study24 to minimize confounding by cytogenetic alterations.
Because CXXC5 is a potential downstream target of GATA2, diagnostic material from 17 cases with GATA2-mutated AML was obtained from the National Cancer Research Institute AML trials tissue bank at University College London and the MLL Munich Leukemia Laboratory. Genomic DNA for mutational analyses was available for 84 cases from the UK MRC AML15 trial; pyrosequencing of the CXXC5 promoter was conducted in 46 cases.
Informed consent was provided by all patients and donors according to the Declaration of Helsinki and subject to ethical board approval.
Cell lines and treatment
Leukemic cell lines K562, KG1a, HL60, NB4, U937, and OCI-AML3, as well as 293T cells were obtained from the DSMZ (Braunschweig, Germany) and cultured as recommended.
Cells were treated with 5-Aza-2′-deoxycytidine (DAC, in dimethyl sulfoxide; Sigma-Aldrich, St Louis, MO) in 12-well plates for 96 hours with 0.2 µM DAC replaced every 24 hours. RNA and DNA were extracted using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany).
For DNA damage experiments, cells were γ-irradiated (4 Gy) in full medium at room temperature.
DNA constructs and transfection
Human CXXC5 cDNA was inserted into a pEGFP-C2 expression vector (Clontech, Mountain View, CA) and transfected (see supplemental Material).
shRNA lentiviral infection
Lentiviral shRNA vectors targeting CXXC5 (pLKO.1; clones TRCN0000144558 and TRCN0000142729) were obtained from Open Biosystems (Thermo Fisher, Lafayette, CO). Cells were infected with viral particles overnight and selected by puromycin treatment (see supplemental Material).
Wnt reporter assay
K562 and KG1a cells were cotransfected with pEGFP-C2-CXXC5/empty pEGFP-C2, M50 Super 8xTOPFlash vector, and pRL-TK vector (see supplemental Material). After 5 hours, cells were seeded in 12-well plates in serum-free medium and incubated overnight. Cells were then treated with 100 ng/mL recombinant Wnt3a protein (R&D Systems, Minneapolis, MN), and luciferase activity was measured in triplicates using the Dual-Luciferase Reporter Assay (Promega).
Western blot
Western blot analyses were performed following standard protocols. Antibodies and incubation conditions are listed in the supplemental Material.
Immunofluorescence
Cells were transferred onto a microscope slide by cytospin centrifugation, fixed in 4% paraformaldehyde, and permeabilized in 0.5% Triton X-100. Staining was performed under standard conditions using antibodies listed in supplemental Table 1. 4′,6-diamidino-2-phenylindole was added for nuclear staining. Confocal microscopy was performed on a Nikon A1Rsi confocal microscope using NIS Elements C software 3.2 (Nikon Instruments, Amsterdam, The Netherlands).
RT-qPCR
RNA was extracted from mononuclear cells using TRIzol reagent (Invitrogen, Carlsbad, CA) and cDNA synthesized with random hexamers and ThermoScript reverse transcriptase (Invitrogen). RT-qPCR was conducted as previously described25 with the TaqMan Universal PCR Master Mix on the ABI 7900HT platform (Applied Biosystems, Warrington, United Kingdom), with normalization of CXXC5 expression to ABL, using optimized assays for analysis of leukemia samples established in the Europe Against Cancer program,26 after validating against B2M and GUS housekeeping genes (see supplemental Material). Relative expression of genes was determined with the comparative cycle threshold (Ct) method,25 calculating the mean of cycle number difference to the control gene, expressed as 2∆Ct. RNA from Jurkat cells was measured in each run as a calibrator. Primers and probes are shown in supplemental Table 1.
Cell-cycle analyses
Cells were fixed in 70% ethanol at 4°C overnight, stained with 50 µg/mL propidium iodide and 100 µg/mL RNAse for 30 minutes at 37°C, and immediately analyzed by flow cytometry using FlowJo software 8.7 (Tree Star, Ashland, OR).
Microarray studies
A microarray-based CXXC5-associated gene-expression profile (GEP) was generated from the MILE and HOVON-SAKK cohorts (discussed before) using a HG-U133 Plus 2.0 Affymetrix platform (Santa Clara, CA; see supplemental Material). Samples were grouped into quartiles (Q1-Q4) according to the median of the 3 CXXC5 probe sets (222996_s_at, 224516_s_at, 233955_x_at) and patients in the lowest and the highest quartile were compared (Q1 vs Q4).
GEP analyses on Wnt-stimulated K562 cells were performed using HG-U133A 2.0 arrays (Affymetrix) as described in the supplemental Material.
Mutational profiling and pyrosequencing
Mutational analyses of CXXC5 were performed as described in the supplemental Material. To investigate the relationship between CXXC5 expression and methylation status, bisulfite conversion of genomic DNA was performed with the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) and subjected to pyrosequencing (see supplemental Material). To investigate whether CXXC5 methylation status correlated with the mutational profile, targeted sequencing was performed using a custom 51-gene panel (HaloPlex Target Enrichment, Agilent Technologies, Santa Clara, CA) on the Illumina HiSeq 2500 platform (see supplemental Material).
Outcome analyses
Outcome analyses were performed on microarray expression data of the HOVON/SAKK cohort (data available for 498 patients, excluding APL) with a median follow-up of 3.4 years. Patients were divided into lower and higher CXXC5-expression groups according to the median of the 3 CXXC5 probe sets (discussed before). Clinical end points and statistical calculations using SPSS software 18.0 (SPSS, Chicago, IL) are described in the supplemental Material.
Results
CXXC5 expression in AML and association with clinical and molecular characteristics
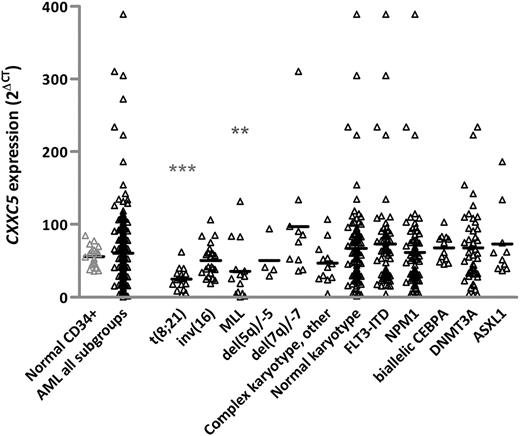
CXXC5 mRNA expression was determined by RT-qPCR in 231 AML patients from the HOVON/SAKK cohort and 19 normal CD34+ cells (Figure 1). Median CXXC5 expression of the total AML cohort did not significantly differ from median expression in normal CD34+ cells. However, AML with MLL rearrangements and t(8;21) had significantly lower CXXC5 expression compared with CD34+ cells (Figure 1). We did not have sufficient del(5q) cases to show downregulation of CXXC5 in del(5q) AML as previously reported.17 However, analysis of 14 selected del(5q)/-5 cases from the MRC AML17 trial confirmed significant downregulation of CXXC5 compared with CD34+ cells (P < .001). AML with CXXC5 expression of higher-than-normal CD34+ cells could not be assigned to any specific subgroup.
CXXC5 mRNA expression in AML and control samples.CXXC5 expression was determined in 231 HOVON/SAKK AML cases and 19 normal CD34+ by RT-qPCR, calculating the mean cycle threshold difference (∆CT) to ABL, expressed as 2∆Ct. Jurkat RNA was used as a calibrator between runs. Horizontal lines indicate median expression values. CXXC5 expression in cases with t(8;21) and MLL rearrangements was significantly lower compared with CD34+ controls. The other subgroups, including cases with biallelic mutations of CEBPA or mutations involving NPM1, DNMT3A, and ASXL1 genes, showed no significant difference in CXXC5 expression compared with CD34+ controls. “Complex karyotype other” indicates cases with complex karyotype without del(5q) or del(7q). **P ≤ .01, ***P ≤ .001 by the Mann-Whitney U test.
CXXC5 mRNA expression in AML and control samples.CXXC5 expression was determined in 231 HOVON/SAKK AML cases and 19 normal CD34+ by RT-qPCR, calculating the mean cycle threshold difference (∆CT) to ABL, expressed as 2∆Ct. Jurkat RNA was used as a calibrator between runs. Horizontal lines indicate median expression values. CXXC5 expression in cases with t(8;21) and MLL rearrangements was significantly lower compared with CD34+ controls. The other subgroups, including cases with biallelic mutations of CEBPA or mutations involving NPM1, DNMT3A, and ASXL1 genes, showed no significant difference in CXXC5 expression compared with CD34+ controls. “Complex karyotype other” indicates cases with complex karyotype without del(5q) or del(7q). **P ≤ .01, ***P ≤ .001 by the Mann-Whitney U test.
Association of microarray-based CXXC5 expression from the HOVON/SAKK cohort (n = 529) with clinical and molecular markers is shown in Table 1. Microarray-based CXXC5 expression correlated well with RT-qPCR results (r = 0.7; P < .001). AML patients were grouped into lower and higher CXXC5 expressers according to the median expression level of all 3 CXXC5 probe sets. Patients with lower CXXC5 expression had a higher incidence of MLL rearrangements (9% vs 4%; P = .03) and t(8;21) (13% vs 2%; P < .001) and a lower incidence of FLT3-ITD (22% vs 32%; P = .01). No significant association was seen with respect to del(5q), likely because of low patient numbers (Table 1). We found correlation of CXXC5 expression with expression of MN1 (Pearson coefficient, r = 0.2; P < .001), WT1 (r = 0.5; P < .001), and ERG (r = 0.3; P < .001). No significant association was observed with respect to mutations of CEBPA, NPM1, IDH1/2, DNMT3A, or ASXL1, nor with EVI1 and BAALC expression (data not shown). Median CXXC5 expression was significantly higher in cases with WT1 mutation (P = .03). There was no association of CXXC5 expression with age, white blood cell count, or sex (Table 1).
Clinical and molecular characteristics according to CXXC5 expression in AML in the HOVON/SAKK cohort
| Characteristic . | CXXC5 lower (n = 264) . | CXXC5 higher (n = 265) . | P . |
|---|---|---|---|
| Age, y | .64 | ||
| Median | 46 | 47 | |
| Range | 15-77 | 15-77 | |
| Sex, % | |||
| Male | 50 | 50 | 1.0 |
| WBC, ×109/L | .89 | ||
| Median | 28 | 29 | |
| Range | 0.3-510 | 0.8-274 | |
| Cytogenetics, %* | |||
| t(8;21); RUNX1-RUNX1T1 | 13 | 2 | <.001 |
| inv(16) or t(16;16); CBFB-MYH11 | 10 | 6 | .15 |
| 11q23 | 9 | 4 | .03 |
| t(6;9); DEK-NUP214 | 1 | 1 | 1.0 |
| inv(3) or t(3;3); RPN1-EVI1 | 1 | 3 | .14 |
| del(5q) | 1 | 1 | .68 |
| Other adverse | 9 | 12 | .57 |
| Normal karyotype | 37 | 48 | .02 |
| Molecular genetics, %† | |||
| FLT3-ITD | 22 | 32 | .01 |
| NPM1 mutated | 30 | 31 | .92 |
| Characteristic . | CXXC5 lower (n = 264) . | CXXC5 higher (n = 265) . | P . |
|---|---|---|---|
| Age, y | .64 | ||
| Median | 46 | 47 | |
| Range | 15-77 | 15-77 | |
| Sex, % | |||
| Male | 50 | 50 | 1.0 |
| WBC, ×109/L | .89 | ||
| Median | 28 | 29 | |
| Range | 0.3-510 | 0.8-274 | |
| Cytogenetics, %* | |||
| t(8;21); RUNX1-RUNX1T1 | 13 | 2 | <.001 |
| inv(16) or t(16;16); CBFB-MYH11 | 10 | 6 | .15 |
| 11q23 | 9 | 4 | .03 |
| t(6;9); DEK-NUP214 | 1 | 1 | 1.0 |
| inv(3) or t(3;3); RPN1-EVI1 | 1 | 3 | .14 |
| del(5q) | 1 | 1 | .68 |
| Other adverse | 9 | 12 | .57 |
| Normal karyotype | 37 | 48 | .02 |
| Molecular genetics, %† | |||
| FLT3-ITD | 22 | 32 | .01 |
| NPM1 mutated | 30 | 31 | .92 |
WBC, white blood cell.
N = 510.
N = 522.
CXXC5-associated GEP in AML patients
In NK-AML (MILE data set, n = 351), we found 659 probe sets comprising 457 genes to be differentially expressed in cases with lower vs higher CXXC5 expression (Q1 vs Q4; see Patients and methods), 295 being upregulated and 162 downregulated (supplemental Table 2 and Figure 1). Among the upregulated genes in the lower CXXC5 expression group were the antiapoptotic proto-oncogenes Survivin and BCL6, as well as genes of aurora kinase signaling (AURKA, DLGAP5, BUB1) and cell-cycling (CCNB1/2, CCNA2, CDK1, CDC20, KIF23, NEK2, UBE2C). Downregulated genes in patients with lower CXXC5 expression included genes that have previously been related to leukemogenesis and outcome in AML (WT1,27 GATA2,28,29 MLL,9 DNMT3B,10 RUNX1,30 MN1,31 MIR155HG,32 MSI233 ). To validate differentially expressed genes from the MILE dataset, we also analyzed CXXC5-associated gene expression in the unselected HOVON/SAKK cohort (supplemental Table 3). In total, 117 of the 295 upregulated genes and 49 of the 162 downregulated genes were confirmed in the HOVON/SAKK cohort, including WT1, GATA2, DNMT3B, RUNX1, and MSI2 (supplemental Figure 2).
Association of CXXC5 and GATA2
The transcription factor GATA2, codownregulated in our GEP analysis, binds the CXXC5 promoter according to ENCODE data (UCSC genome browser; http://genome.ucsc.edu), suggesting regulation of CXXC5 by GATA2. To further investigate this potential interaction, we overexpressed GATA2 in 293T cells and found upregulation of CXXC5 protein compared with vector controls (supplemental Figure 3), which was confirmed at the mRNA level (mean 2.8-fold increase of CXXC5 expression compared with control; data not shown), supporting a functional interplay of both genes. We next measured CXXC5 mRNA levels in 17 AML patients with GATA2 mutations and showed a significantly lower CXXC5 expression than in normal CD34+ cells (median, 15 [range, 5-39]; P < .0001).
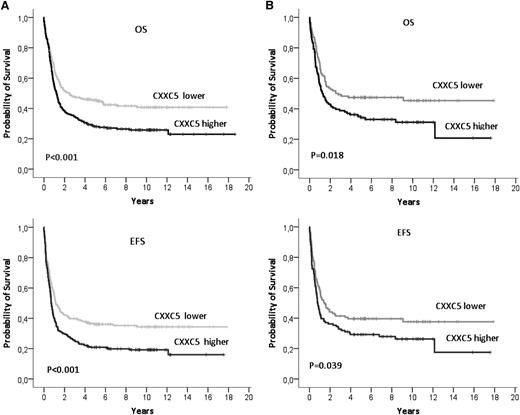
CXXC5 expression and outcome in AML
In 498 patients from the HOVON/SAKK cohort (APL excluded) analyzed for outcome, patients with lower CXXC5 expression (below the median expression level) had a trend to a higher complete remission (CR) rate (84% vs 77%; P = .07) and a lower relapse rate compared with patients with higher CXXC5 expression (45% vs 59% at 5 years; P = .007). Overall survival (OS) and event-free survival (EFS) were significantly better in lower compared with higher CXXC5-expressing AML cases (5-year OS: 95% confidence interval [CI], 46% [40%-52%] vs 28% [22%-34%]; P < .001; 5-year EFS: 36% [30%-42%] vs 21% [16%-26%]; P < .001; Figure 2A). The prognostic significance of CXXC5 was independent of known clinical and molecular risk factors in multivariate analyses (Table 2). There was no significant heterogeneity in the impact of CXXC5 on outcome between AML risk groups (supplemental Figure 4). CXXC5 was also significant for OS and EFS when treated as a continuous variable (data not shown). A significant impact of CXXC5 expression on OS and EFS was confirmed in the cohort of 231 AML patients using RT-qPCR median expression values (Figure 2B).
Overall survival (OS) and event-free survival (EFS) in HOVON/SAKK AML patients according to CXXC5 expression. (A) Survival according to microarray-based CXXC5 expression (n = 498). CXXC5 expression below the median (lower): median OS of 30 months (95% CI, 4-57); CXXC5 expression above the median (higher): median OS of 15 months (95% CI, 12-18). CXXC5 lower: median EFS of 14 months (95% CI, 10-18); CXXC5 higher: median EFS of 9 months (95% CI, 7-10). (B) Survival according to RT-qPCR expression (n = 231). Lower and higher CXXC5 expression groups were defined according to the median CXXC5 expression value, determined by the mean cycle threshold difference (∆CT) to ABL, expressed as 2∆Ct. Jurkat RNA was used to calibrate between runs. CXXC5 was more highly expressed than ABL, with a ∆CT of 2.3 at the median CXXC5 level.
Overall survival (OS) and event-free survival (EFS) in HOVON/SAKK AML patients according to CXXC5 expression. (A) Survival according to microarray-based CXXC5 expression (n = 498). CXXC5 expression below the median (lower): median OS of 30 months (95% CI, 4-57); CXXC5 expression above the median (higher): median OS of 15 months (95% CI, 12-18). CXXC5 lower: median EFS of 14 months (95% CI, 10-18); CXXC5 higher: median EFS of 9 months (95% CI, 7-10). (B) Survival according to RT-qPCR expression (n = 231). Lower and higher CXXC5 expression groups were defined according to the median CXXC5 expression value, determined by the mean cycle threshold difference (∆CT) to ABL, expressed as 2∆Ct. Jurkat RNA was used to calibrate between runs. CXXC5 was more highly expressed than ABL, with a ∆CT of 2.3 at the median CXXC5 level.
Multivariate analyses for CXXC5 expression
| Variable* . | OR/HR . | 95% CI . | P . |
|---|---|---|---|
| Relapse rate | |||
| Low CXXC5† | 0.5 | 0.3-0.8 | .007 |
| Cytogenetic risk | 1.9 | 1.2-2.9 | .005 |
| WBC | 1.06 | 1.0-1.1 | .005 |
| CEBPA mut., biallelic | 0.4 | 0.2-1.0 | .05 |
| ASXL1 mut. | 9.5 | 2.0-45 | .004 |
| Overall survival | |||
| Low CXXC5† | 0.6 | 0.5-0.8 | .001 |
| Cytogenetic risk | 1.7 | 1.3-2.1 | <.001 |
| Age | 1.2 | 1.0-1.3 | .004 |
| FLT3-ITD/NPM1 wt | 1.7 | 1.2-2.5 | .004 |
| NPM1 mut./FLT3-ITD neg. | 0.6 | 0.4-0.8 | .006 |
| CEBPA mut., biallelic | 0.3 | 0.2-0.6 | <.001 |
| DNMT3A mut. | 1.4 | 1.1-2.0 | .02 |
| ASXL1 mut. | 1.6 | 1.0-2.5 | .04 |
| Event-free survival | |||
| Low CXXC5† | 0.6 | 0.5-0.8 | <.001 |
| Cytogenetic risk | 1.6 | 1.3-2.0 | <.001 |
| WBC | 1.02 | 1.0-1.0 | .04 |
| FLT3-ITD/NPM1 wt | 1.7 | 1.2-2.5 | .006 |
| NPM1 mut./FLT3-ITD neg. | 0.6 | 0.4-0.9 | .009 |
| CEBPA mut., biallelic | 0.3 | 0.2-0.6 | <.001 |
| DNMT3A mut. | 1.4 | 1.0-1.8 | .04 |
| ASXL1 mut. | 2.0 | 1.2-3.2 | .005 |
| Variable* . | OR/HR . | 95% CI . | P . |
|---|---|---|---|
| Relapse rate | |||
| Low CXXC5† | 0.5 | 0.3-0.8 | .007 |
| Cytogenetic risk | 1.9 | 1.2-2.9 | .005 |
| WBC | 1.06 | 1.0-1.1 | .005 |
| CEBPA mut., biallelic | 0.4 | 0.2-1.0 | .05 |
| ASXL1 mut. | 9.5 | 2.0-45 | .004 |
| Overall survival | |||
| Low CXXC5† | 0.6 | 0.5-0.8 | .001 |
| Cytogenetic risk | 1.7 | 1.3-2.1 | <.001 |
| Age | 1.2 | 1.0-1.3 | .004 |
| FLT3-ITD/NPM1 wt | 1.7 | 1.2-2.5 | .004 |
| NPM1 mut./FLT3-ITD neg. | 0.6 | 0.4-0.8 | .006 |
| CEBPA mut., biallelic | 0.3 | 0.2-0.6 | <.001 |
| DNMT3A mut. | 1.4 | 1.1-2.0 | .02 |
| ASXL1 mut. | 1.6 | 1.0-2.5 | .04 |
| Event-free survival | |||
| Low CXXC5† | 0.6 | 0.5-0.8 | <.001 |
| Cytogenetic risk | 1.6 | 1.3-2.0 | <.001 |
| WBC | 1.02 | 1.0-1.0 | .04 |
| FLT3-ITD/NPM1 wt | 1.7 | 1.2-2.5 | .006 |
| NPM1 mut./FLT3-ITD neg. | 0.6 | 0.4-0.9 | .009 |
| CEBPA mut., biallelic | 0.3 | 0.2-0.6 | <.001 |
| DNMT3A mut. | 1.4 | 1.0-1.8 | .04 |
| ASXL1 mut. | 2.0 | 1.2-3.2 | .005 |
CI, confidence interval; HR, hazard ratio; mut., mutated; neg., negative; OR, odds ratio; WBC, white blood cell count; wt, wild-type.
Variables considered for model inclusion were (see supplemental Material for details): CXXC5 expression (below vs above median level), cytogenetic risk groups (good, intermediate, unfavorable), age (10-year increase), WBC (10/nl increase), FLT3-ITD34 /NPM1 wild-type (present vs absent), NPM1 mutation35 /FLT3-ITD negative (present vs absent), CEBPA biallelic mutation36 (present vs absent), DNMT3A mutation37 (present vs absent), ASXL1 mutation38 (present vs absent), WT1 mutation (present vs absent), WT1 expression (lower vs higher than median).
Below median expression level.
CXXC5 impairs the canonical Wnt pathway
Because CXXC5 is known to inhibit the canonical Wnt pathway in nontumorous cells, we determined its impact on Wnt signaling in leukemic cell lines. Overexpression of CXXC5 in K562 and KG1a cells attenuated upregulation of total β-catenin and active β-catenin 5 hours after Wnt3a stimulation compared with vector controls (Figure 3A). Moreover, upregulation of Wnt target genes Axin2 and Survivin by Wnt3a was significantly decreased after expression of CXXC5 in both cell lines as determined by RT-qPCR (Figure 3B). In addition, we evaluated differences in Wnt activation by Wnt reporter assay. CXXC5 expression resulted in decreased luciferase activity after Wnt3a stimulation in K562 and KG1a compared with vector controls (Figure 3C).
CXXC5 attenuates Wnt signaling in leukemic cells. Cells overexpressing EGFP-tagged CXXC5 or stably infected with lentiviral CXXC5 shRNA were treated with 100 ng/mL recombinant Wnt3a. (A) Decreased upregulation of total β-catenin and active β-catenin 5 hours after Wnt3a treatment in CXXC5-overexpressing cells compared with vector controls (Western blot analysis). A representative figure is shown (transfection efficiency of 59% for K562 and 42% for KG1a by flow cytometry). (B) Decreased upregulation of Wnt target genes Axin2 and Survivin 10 hours after Wnt3a treatment in CXXC5-overexpressing cells compared with vector controls as determined by RT-qPCR. Mean values of 2 independent experiments with standard deviations are shown (K562 Axin2: 1.4-fold vs 3.1-fold; K562 Survivin: 0.9-fold vs 1.2-fold; KG1a Axin2: 0.7-fold vs 1.7-fold; KG1a Survivin: 0.6-fold vs 1.3-fold; both P ≤ .05 by the Mann-Whitney U test[*]). (C) Impaired Wnt activation as determined by Wnt reporter assay activity 5 hours after Wnt3a treatment in CXXC5-overexpressing cells compared with vector controls. Mean values of 2 independent experiments with standard deviations are shown (K562: 4.7-fold vs 19.9-fold; KG1a: 0.9-fold vs 2.8-fold; *P ≤ .05 and **P ≤ .01 by the Mann-Whitney U test). (D) Impaired cell proliferation after CXXC5 overexpression (i, iii) and increased proliferation after CXXC5 knockdown (ii, iv) as determined by cell count (trypan blue staining; iii-iv) and MTT assay (i-ii) 24, 48, and 72 hours after start of treatment (days 1-3). Wnt3a protein was replaced every 24 hours. Mean values of 2 independent experiments performed in quadruplicates with standard deviations are shown. Co, vector control; scr, scrambled shRNA control; shR, CXXC5 shRNA. P values are shown in the figure (**P ≤ .01 and ***P ≤ .001 by the Mann-Whitney U test).
CXXC5 attenuates Wnt signaling in leukemic cells. Cells overexpressing EGFP-tagged CXXC5 or stably infected with lentiviral CXXC5 shRNA were treated with 100 ng/mL recombinant Wnt3a. (A) Decreased upregulation of total β-catenin and active β-catenin 5 hours after Wnt3a treatment in CXXC5-overexpressing cells compared with vector controls (Western blot analysis). A representative figure is shown (transfection efficiency of 59% for K562 and 42% for KG1a by flow cytometry). (B) Decreased upregulation of Wnt target genes Axin2 and Survivin 10 hours after Wnt3a treatment in CXXC5-overexpressing cells compared with vector controls as determined by RT-qPCR. Mean values of 2 independent experiments with standard deviations are shown (K562 Axin2: 1.4-fold vs 3.1-fold; K562 Survivin: 0.9-fold vs 1.2-fold; KG1a Axin2: 0.7-fold vs 1.7-fold; KG1a Survivin: 0.6-fold vs 1.3-fold; both P ≤ .05 by the Mann-Whitney U test[*]). (C) Impaired Wnt activation as determined by Wnt reporter assay activity 5 hours after Wnt3a treatment in CXXC5-overexpressing cells compared with vector controls. Mean values of 2 independent experiments with standard deviations are shown (K562: 4.7-fold vs 19.9-fold; KG1a: 0.9-fold vs 2.8-fold; *P ≤ .05 and **P ≤ .01 by the Mann-Whitney U test). (D) Impaired cell proliferation after CXXC5 overexpression (i, iii) and increased proliferation after CXXC5 knockdown (ii, iv) as determined by cell count (trypan blue staining; iii-iv) and MTT assay (i-ii) 24, 48, and 72 hours after start of treatment (days 1-3). Wnt3a protein was replaced every 24 hours. Mean values of 2 independent experiments performed in quadruplicates with standard deviations are shown. Co, vector control; scr, scrambled shRNA control; shR, CXXC5 shRNA. P values are shown in the figure (**P ≤ .01 and ***P ≤ .001 by the Mann-Whitney U test).
Overexpression of CXXC5 led to a significant decrease in proliferation of K562, KG1a, and HL60 cells treated with Wnt3a protein as determined by cell count and MTT assay (Figure 3D). This was accompanied by increased apoptosis measured by Annexin V staining (data not shown). Accordingly, knockdown of CXXC5 (supplemental Figure 5) resulted in increased proliferation of Wnt-stimulated cells (Figure 3D). Without concomitant Wnt3a treatment, cell proliferation was only moderately decreased in K562 and HL60 cells overexpressing CXXC5, but was not significantly changed in KG1a or in any of the CXXC5 knockdown cells (data not shown).
Previous studies suggested that CXXC5 interacts with disheveled (Dvl) proteins as a mechanism of Wnt inhibition.4,5 However, we did not observe colocalization of EGFP-tagged CXXC5 with endogenous Dvl1, Dvl2, or Dvl3 in confocal microscopy or binding in immunoprecipitation experiments in K562 (data not shown).
To gain further insights into the cellular pathways involved in the interaction of CXXC5 with Wnt signaling, we conducted microarray-based GEP on Wnt-stimulated K562 cells after overexpression and knockdown of CXXC5, respectively (supplemental Table 4). We found a substantial overlap of differentially expressed genes in both models: 77 of 186 upregulated genes in CXXC5-overexpressing cells showed corresponding downregulation in knockdown cells. Similarly, 65 of 137 downregulated genes in CXXC5-overexpressing cells were upregulated in CXXC5 knockdown cells. Several signaling pathways were significantly enriched in both CXXC5-overexpressing and knockdown cells, such as TRAIL, c-MYC, mTOR, and IGF1 (supplemental Figure 6). We found Wnt target genes Survivin, TPM1, SRSF3, MET, HES1, and FSCN1 to be upregulated in CXXC5 knockdown cells, indicating activated Wnt signaling. HES1 expression was accordingly downregulated in CXXC5-overexpressing cells. Wnt repressors HBP1 and AXIN1 were upregulated in cells overexpressing CXXC5 (1.6-fold [P = .001] and 1.2-fold [P ≤ .0001]) and accordingly downregulated in knockdown cells (1.7-fold [P = .001] and 1.4-fold [P = .003]). In addition, CXXC5-knockdown cells showed significant upregulation of Wnt activators CAV1 and APPL1 (supplemental Table 4).
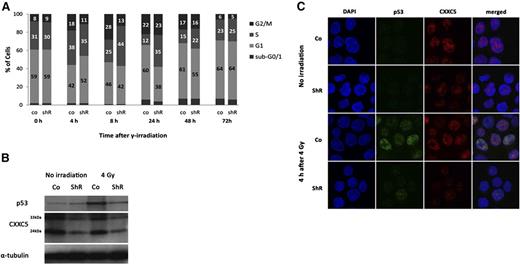
Knockdown of CXXC5 reduces p53 activation and cell-cycle arrest upon DNA damage
Because CXXC5 has been shown to affect p53 signaling,3 we performed cell-cycle analyses after γ-irradiation of OCI-AML3 cells (harboring wild-type p53) to see whether knockdown of CXXC5 alters the p53-dependent DNA damage response in leukemic cells. OCI-AML3 stably infected with lentiviral CXXC5 shRNA clones TRCN0000144558 and TRCN0000142729 showed a reduced and delayed G1 and a reduced G2 arrest after irradiation with 4 Gy (Figure 4A). In addition, irradiation-induced upregulation of p53 protein was decreased after CXXC5 knockdown compared with controls (Figure 4B). This was accompanied by impaired nuclear translocation of p53 (Figure 4C). Knockdown of CXXC5 in K562 (p53 deficient) did not alter cell-cycle distribution after irradiation (data not shown), further underlining the p53-dependent impact of CXXC5 on DNA-damage response in leukemic cells.
Inhibition of DNA damage–induced cell-cycle arrest and p53 activation after knockdown of CXXC5 in OCI-AML3 cells. Cells were stably infected with lentiviral CXXC5 shRNA (shR) or scrambled control (co) and irradiated with 4 Gy. (A) Reduced and delayed G1 and G2 arrest after irradiation compared with controls. A representative figure is shown. Numbers in bars correspond to percentages of cells. (B) Decreased upregulation of p53 protein 4 hours after irradiation compared with control as determined by Western blot. A representative blot is shown. The 2 bands for CXXC5 correspond to the 24-kDa and 33-kDa isoforms. (C) Decreased upregulation and nuclear localization of p53 protein 4 hours after irradiation compared with control in confocal microscopy. Representative images are shown. Original magnification ×60 for all images.
Inhibition of DNA damage–induced cell-cycle arrest and p53 activation after knockdown of CXXC5 in OCI-AML3 cells. Cells were stably infected with lentiviral CXXC5 shRNA (shR) or scrambled control (co) and irradiated with 4 Gy. (A) Reduced and delayed G1 and G2 arrest after irradiation compared with controls. A representative figure is shown. Numbers in bars correspond to percentages of cells. (B) Decreased upregulation of p53 protein 4 hours after irradiation compared with control as determined by Western blot. A representative blot is shown. The 2 bands for CXXC5 correspond to the 24-kDa and 33-kDa isoforms. (C) Decreased upregulation and nuclear localization of p53 protein 4 hours after irradiation compared with control in confocal microscopy. Representative images are shown. Original magnification ×60 for all images.
Mutational analyses of CXXC5
We performed DNA sequencing of CXXC5 in 84 AML patients to investigate the presence of CXXC5 mutations. No CXXC5 mutations were found, suggesting that deregulation of CXXC5 by gene mutations plays no major role in AML.
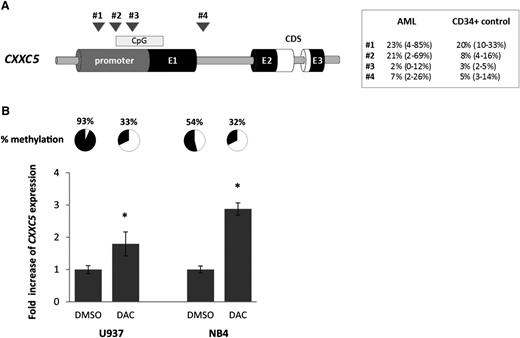
Association of CXXC5 expression with promoter methylation
To evaluate epigenetic silencing as a mechanism of CXXC5 downregulation in AML, we conducted pyrosequencing in 46 AML samples and 6 normal CD34+ samples at 4 different regions of the CXXC5 promoter (#1-3) and intron 1 (#4; Figure 5A). In AML, the degree of methylation of region #2 was inversely correlated with CXXC5 mRNA expression levels (r = −0.32; P = .03). A higher degree of methylation at regions #1 and #2 was observed in cases harboring IDH1/2 mutations (median, 42% vs 15%; P = .004; and median, 29% vs 12%; P = .05, respectively), whereas methylation at region #2 was lower in cases with DNMT3A mutations (9% vs 20%; P = .05). There was no correlation between mutation status and gene expression, although sample numbers were small. No relationship was observed between the mutational status of other genes in the panel and methylation, including WT1, TET2, and ASXL1 (see supplemental Figure 7).
Epigenetic regulation of CXXC5. (A) Schematic figure of methylation analysis by pyrosequencing within the CXXC5 gene and median methylation values (with range) for AML samples (n = 46) and healthy controls (normal BM CD34+ cells, n = 6). CDS, coding region; CpG, CpG island; E1-3, 3 CXXC5 exons. Four different regions in the CXXC5 promoter (#1-3) and intron 1 (#4) were analyzed: (#1) −1343 to −1213 (2 CpGs); (#2) −914 to −741 (3 CpGs); (#3) −749 to −593 (5 CpGs); (#4) +1955 to +2179 (4 CpGs). The previous study of Treppendahl et al17 investigated a site further upstream in the CXXC5 promoter (−20 440 to −20 330, personal communication Marianne Treppendahl, Department of Hematology, Rigshospitalet Copenhagen, Denmark). Methylation of region #2 was significantly higher in AML than in healthy controls (P = .04); methylation of region #3 was significantly lower (P = .001). No difference between AML and controls was observed in the other 2 regions. P values were calculated by the Mann-Whitney U test. (B) Upregulation of CXXC5 after demethylation of region #2. U937 and NB4 cells were treated with 5-Aza-2′-deoxycytidine (DAC) for 96 hours and CXXC5 expression was determined by RT-qPCR. Values are normalized to dimethyl sulfoxide control. Mean values of 2 independent experiments with standard deviations are shown (*P ≤ .05 by the Mann-Whitney U test). Demethylation of region #2 was confirmed by pyrosequencing and mean methylation percentages of 2 independent experiments are shown.
Epigenetic regulation of CXXC5. (A) Schematic figure of methylation analysis by pyrosequencing within the CXXC5 gene and median methylation values (with range) for AML samples (n = 46) and healthy controls (normal BM CD34+ cells, n = 6). CDS, coding region; CpG, CpG island; E1-3, 3 CXXC5 exons. Four different regions in the CXXC5 promoter (#1-3) and intron 1 (#4) were analyzed: (#1) −1343 to −1213 (2 CpGs); (#2) −914 to −741 (3 CpGs); (#3) −749 to −593 (5 CpGs); (#4) +1955 to +2179 (4 CpGs). The previous study of Treppendahl et al17 investigated a site further upstream in the CXXC5 promoter (−20 440 to −20 330, personal communication Marianne Treppendahl, Department of Hematology, Rigshospitalet Copenhagen, Denmark). Methylation of region #2 was significantly higher in AML than in healthy controls (P = .04); methylation of region #3 was significantly lower (P = .001). No difference between AML and controls was observed in the other 2 regions. P values were calculated by the Mann-Whitney U test. (B) Upregulation of CXXC5 after demethylation of region #2. U937 and NB4 cells were treated with 5-Aza-2′-deoxycytidine (DAC) for 96 hours and CXXC5 expression was determined by RT-qPCR. Values are normalized to dimethyl sulfoxide control. Mean values of 2 independent experiments with standard deviations are shown (*P ≤ .05 by the Mann-Whitney U test). Demethylation of region #2 was confirmed by pyrosequencing and mean methylation percentages of 2 independent experiments are shown.
We tested methylation of region #2 in different leukemic cell lines. KG1a, K562, and HEL cells were unmethylated (<3%), whereas Kasumi-1 (24%), NB4 (68%), and U937 (95%) showed a high degree of methylation.
To investigate epigenetic regulation of CXXC5 at region #2, we treated the highly methylated cell lines U937 and NB4 with 0.2 µM DAC. After 96 hours, mean methylation at region #2 was markedly decreased and CXXC5 expression was increased 1.8-fold and 2.9-fold in U937 and NB4, respectively (both P < .05; Figure 5B), suggesting silencing of CXXC5 by promoter hypermethylation in AML. This was also reflected at the protein level (supplemental Figure 8). No change in CXXC5 expression was seen when cells were treated with 1 to 50 nM cytarabine and no effect was observed in unmethylated KG1a and K562 cells (data not shown).
Discussion
Identification of prognostically relevant leukemic pathways in AML provides novel insights into disease pathogenesis and may inform targeted therapy approaches. Aberrant CXXC5 expression has recently been described in a small cohort of AML, and preliminary data suggested an impact of CXXC5 expression on outcome.21 Here, we found evidence for a tumor-suppressor function of CXXC5 in AML and demonstrated its independent prognostic significance.
Investigating CXXC5 expression in the HOVON/SAKK multicenter AML trial cohort, we showed that CXXC5 was significantly downregulated in AML with MLL rearrangements and t(8;21). Downregulation of CXXC5 has previously been reported in 22 AML/high-risk MDS patients with del(5q),17 in accordance with our experimental findings.
GEP analyses further revealed that CXXC5 expression was codownregulated with genes for which inactivating mutations or silencing of the wild-type allele/protein have been implicated in leukemogenesis and cancer development, such as GATA2,28,29 MLL,39 DNMT3B,40 and RUNX1,30,41 which is in line with a recent study investigating a single-center cohort of 48 AML cases.42 We investigated 17 AML samples with GATA2 mutations and found CXXC5 expression to be significantly downregulated. We further demonstrated that overexpression of GATA2 leads to upregulation of CXXC5, suggesting transcriptional regulation of CXXC5 by GATA2, which is in line with published ENCODE data. Thus, low expression of CXXC5 might be a common downstream effect of different leukemic pathways, including GATA2 mutations.
We demonstrated that CXXC5 attenuates Wnt signaling in leukemic cell lines, a crucial pathway for leukemic self-renewal and survival.8 Downregulation of CXXC5 might therefore contribute to leukemogenesis by promoting Wnt signaling. Wnt inhibition by CXXC5 has been described in nontumorous tissue, and interaction of CXXC5 with Dvl proteins was identified as an underlying mechanism.4,5 Here, we did not observe interaction of CXXC5 with Dvl proteins. This might be explained by the fact that in the leukemic cell lines tested, CXXC5 (EGFP-tagged and endogenous protein) was predominantly localized in the nucleus, in accordance with in silico analyses and results in NB4, HeLa, and MCF7 cells.3,20 In contrast, Kim et al and Andersson et al found CXXC5 (and its interaction with Dvl) localized in the cytoplasm.4,5 Thus, interaction of CXXC5 with Dvl proteins and its relevance for Wnt attenuation might strongly depend on the cellular context. Interestingly, Kim et al demonstrated that the ability of CXXC5 to bind Dvl was less important for Wnt inhibition than presence of the CXXC domain, further suggesting other mechanisms of inhibition.4 It has been shown that CXXC5 interacts with the vitamin D receptor,43 which could lead to Wnt suppression. In our GEP analysis of Wnt-stimulated K562 cells, we identified Wnt repressors HBP1 and AXIN1 significantly altered by CXXC5 expression, which could mediate CXXC5-associated effects on Wnt signaling.
Small molecules targeting the Wnt pathway exhibited marked antileukemic activity in vitro,44 and there is recent evidence that Wnt inhibition is particularly effective in leukemic cells with del(5q).45 Therefore, AML patients with low CXXC5 expression might be candidates for Wnt inhibitor treatment.
In MCF7 and HeLa cells containing wild-type p53, CXXC5 has been shown to promote p53 signaling through binding of ATM kinase with its CXXC domain.3 Similarly, in our study, inactivation of CXXC5 impaired p53-dependent cell-cycle arrest after DNA damage. Low CXXC5 expression might thus increase genomic instability in AML. In GEP analysis, several cell-cycle genes (CCNB1/2, CCNA2, CDK1, CDC2, CDC20, KIF23) were upregulated in patients with low CXXC5 expression, further suggesting involvement of CXXC5 in cell-cycle processes in leukemia.
Lower expression of CXXC5 predicted a lower relapse rate and better long-term survival in AML patients. Our results confirm data from Astori et al who performed univariate outcome analyses on 2 German AML trial cohorts using published microarray data.21 In addition, a recent study related overexpression of CXXC5 to inferior OS in breast cancer.46 In our analysis, low expression of CXXC5 was associated with known prognostic factors (higher frequency of MLL rearrangements and t(8;21) and lower frequency of FLT3-ITD), but its prognostic impact remained significant in multivariate analyses after adjustment for these factors. Thus, CXXC5 expression defines a subgroup of AML with distinct outcome independent of other cytogenetic and molecular markers. It is unclear why lower CXXC5 expression with its activating function on Wnt signaling confers a better prognosis, because Wnt activation has been associated with inferior outcome in AML.47 However, it has recently been shown that high expression of LEF1, a key activator of Wnt signaling, is associated with a better prognosis in AML.48 Moreover, other functions of CXXC5 than Wnt regulation may be relevant for prognosis and might increase chemosensitivity.21,42
In accordance with previous studies, we did not find any mutation of CXXC5 in AML. Genetic alterations of CXXC5 seem to be rare, and infrequent, small intragenic deletions as described in a patient with NK-AML,18 or haploinsufficiency due to del(5q), cannot explain the wide range of expression seen in AML. Treppendahl et al found that several cases with del(5q) had <50% CXXC5 expression levels, suggesting additional mechanisms of inactivation than haploinsufficiency.17 WT1 has been shown to regulate CXXC5 expression4 and was indeed strongly correlated with CXXC5 expression in our GEP analysis. In addition, we found evidence for silencing of CXXC5 by promoter hypermethylation. Of note, this was only evident in 1 region of the CpG shore (−914 to −741), not in other regions of the CXXC5 promoter, which could explain negative results of a previous CXXC5 methylation analysis, investigating a site further upstream (−20440 to −20330).17 Thus, epigenetic modification as well as transcriptional regulation by factors like GATA2 and WT1 might contribute to aberrant CXXC5 expression in AML.
Further studies are needed to investigate mechanisms of CXXC5 regulation, the spectrum of molecular interactions of CXXC5, and its role in leukemic pathways, particularly in cases with MLL rearrangements and t(8;21). Recent studies demonstrated that CXXC5 binds DNA and acts as a transcriptional activator as well as a repressor.49 In addition, the CXXC5 paralog CXXC4 was shown to bind unmethylated CpG-rich DNA and interact with TET2, and a similar role was postulated for CXXC5.50
In summary, we have investigated the 5q31 gene CXXC5 in AML with respect to clinical characteristics and prognosis and gene expression and its functional implications. Our data suggest a role of CXXC5 as a tumor suppressor in different AML subgroups. Inactivation of CXXC5 characterizes AML with a better outcome, which might be particularly sensitive to Wnt inhibitor treatment.
These data were presented in part at the 55th American Society of Hematology meeting, New Orleans, December 2013.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the FACS core facility at Guy’s Hospital, London, for support with flow cytometry and the BRC Genomics Laboratory, King’s College London for enabling high-throughput mutational analyses. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health. Immunofluorescence was conducted at the Nikon Imaging Centre at King’s College London.
This study was supported by Deutsche Krebshilfe, Germany (A.K.); the National Institute for Health Research (NIHR) under its Programme Grants for Applied Research Programme (RP- PG-0108-10093); the NIHR Biomedical Research Centre (BRC) based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London; the Guy’s and St. Thomas’ Charity (D.G.); Leukaemia & Lymphoma Research of Great Britain (D.G., E.S., R. Dillon, R.E.G., D.C.L., M.J.); the UCL/UCLH NIHR BRC (R.E.G., D.C.L.); Leukaemia & Lymphoma NI (LLNI) (K.I.M.); and SIRIC grant #INCa-DGOS-Inserm 6038 (E.D.).
Authorship
Contribution: A.K. designed and performed the research, analyzed the data, and wrote the manuscript; P.J.M.V. and M.A.S. provided microarray and outcome data and contributed to the analysis of the study; A.I. contributed to laboratory work; R.K.H. undertook statistical analyses; K.I.M. provided and analyzed microarray data; R.E.G., A.G., S.S., E.D., and D.C.L. characterized and provided patient samples; M.F.K., R. Dillon, and M.J. contributed to laboratory work; T.H. provided microarray data and patient samples; R. Delwel and B.L. provided microarray and outcome data; C.D.B. contributed to the analysis of the study and reviewed the manuscript; E.S. and D.G. supervised the study, contributed to the design and analysis of the study and to the writing of the manuscript; and A.K.B. was chief investigator of the MRC AML15 trial, provided clinical data, and reviewed the manuscript.
Conflict-of-interest disclosure: The MILE study was supported in part by Roche Molecular Systems Inc. T.H. and S.S. are part owners of the MLL Munich Leukemia Laboratory. The remaining authors declare no competing financial interests.
Correspondence: David Grimwade, Department of Medical & Molecular Genetics, 8th Floor, Tower Wing, Guy’s Hospital, London SE1 9RT, United Kingdom; e-mail: david.grimwade@genetics.kcl.ac.uk.

![Figure 3. CXXC5 attenuates Wnt signaling in leukemic cells. Cells overexpressing EGFP-tagged CXXC5 or stably infected with lentiviral CXXC5 shRNA were treated with 100 ng/mL recombinant Wnt3a. (A) Decreased upregulation of total β-catenin and active β-catenin 5 hours after Wnt3a treatment in CXXC5-overexpressing cells compared with vector controls (Western blot analysis). A representative figure is shown (transfection efficiency of 59% for K562 and 42% for KG1a by flow cytometry). (B) Decreased upregulation of Wnt target genes Axin2 and Survivin 10 hours after Wnt3a treatment in CXXC5-overexpressing cells compared with vector controls as determined by RT-qPCR. Mean values of 2 independent experiments with standard deviations are shown (K562 Axin2: 1.4-fold vs 3.1-fold; K562 Survivin: 0.9-fold vs 1.2-fold; KG1a Axin2: 0.7-fold vs 1.7-fold; KG1a Survivin: 0.6-fold vs 1.3-fold; both P ≤ .05 by the Mann-Whitney U test[*]). (C) Impaired Wnt activation as determined by Wnt reporter assay activity 5 hours after Wnt3a treatment in CXXC5-overexpressing cells compared with vector controls. Mean values of 2 independent experiments with standard deviations are shown (K562: 4.7-fold vs 19.9-fold; KG1a: 0.9-fold vs 2.8-fold; *P ≤ .05 and **P ≤ .01 by the Mann-Whitney U test). (D) Impaired cell proliferation after CXXC5 overexpression (i, iii) and increased proliferation after CXXC5 knockdown (ii, iv) as determined by cell count (trypan blue staining; iii-iv) and MTT assay (i-ii) 24, 48, and 72 hours after start of treatment (days 1-3). Wnt3a protein was replaced every 24 hours. Mean values of 2 independent experiments performed in quadruplicates with standard deviations are shown. Co, vector control; scr, scrambled shRNA control; shR, CXXC5 shRNA. P values are shown in the figure (**P ≤ .01 and ***P ≤ .001 by the Mann-Whitney U test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/19/10.1182_blood-2014-12-613703/4/m_2985f3.jpeg?Expires=1769800582&Signature=DCztclO3oZNy3HX55WJNXCRS66T0BngC4zbnjWxWlDPS5PDlbTOzbFDw5LshskwPo7K5Vo7d7~F-0n7xYPyf7xAzebtJhCodKhqpFEcGkHjod9plDjtDubUj~vIvoINcj17J86JRUJEEc5i1PjM48a0pvTQzbYbI1PU72bYtskRv~GXmnXaW9V9Ivx6BeeWCE89lH2LvN0LVSlFV-rRmzOr5IflI7azvCVORaChVVMxf-5NMbdWFu0E1pbx9VgPZHfNz5iMNABz4zFRueSN9x7bWM1cdpPGovpnHBmHcW-2qaK4xC575nHIuZlP3tQ96dFDqOToqIyiOLdDftQMkhw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)