Key Points
Zebrafish idh1 plays an important role in the regulation of myelopoiesis and definitive hematopoiesis.
Expression of human IDH1-R132H and its zebrafish orthologue induced an increase in myelopoiesis and 2-hydroxyglutrate.
Abstract
Isocitrate dehydrogenase 1 mutation (IDH1-R132H) was recently identified in acute myeloid leukemia with normal cytogenetics. The mutant enzyme is thought to convert α-ketoglutarate to the pathogenic 2-hydroxyglutarate (2-HG) that affects DNA methylation via inhibition of ten-eleven translocation 2. However, the role of wild-type IDH1 in normal hematopoiesis and its relevance to acute myeloid leukemia is unknown. Here we showed that zebrafish idh1 (zidh1) knockdown by morpholino and targeted mutagenesis by transcription activator–like effector nuclease might induce blockade in myeloid differentiation, as evident by an increase in pu.1 and decrease in mpo, l-plastin, and mpeg1 expression, and significantly reduce definitive hematopoiesis. Morpholino knockdown of zidh2 also induced a blockade in myeloid differentiation but definitive hematopoiesis was not affected. The hematopoietic phenotype of zidh1 knockdown was not rescuable by zidh2 messenger RNA, suggesting nonredundant functions. Overexpression of human IDH1-R132H or its zebrafish ortholog resulted in 2-HG elevation and expansion of myelopoiesis in zebrafish embryos. A human IDH1-R132H–specific inhibitor (AGI-5198) significantly ameliorated both hematopoietic and 2-HG responses in human but not zebrafish IDH1 mutant expression. The results provided important insights to the role of zidh1 in myelopoiesis and definitive hematopoiesis and of IDH1-R132H in leukemogenesis.
Introduction
Recent advances in next-generation sequencing in acute myeloid leukemia (AML) have resulted in the identification of novel gene mutations with hitherto unknown functions in leukemogenesis.1 In particular, mutations of isocitrate dehydrogenase 1/2 (IDH1/2), enzymes involved in citric acid cycle in intermediary metabolism, were identified in approximately 30% of cytogenetically normal AML, suggesting a pathogenetic link in leukemia initiation.2,3 IDH1 mutation in AML occurs almost exclusively at codon 132 and IDH2 mutations at both codons 140 and 172 and in all of these mutations arginine was substituted.4 IDH mutations confer novel substrate specificity to the enzyme and instead of converting isocitrate to α-ketoglutarate, the latter is converted to 2-hydroxyglutarate (2-HG).5,6 2-HG may act as oncometabolite, resulting in epigenetic alteration, genetic instability, and malignant transformation of hematopoietic cells.7-11 Importantly, raised serum 2-HG in AML patients has been correlated with IDH mutations and is being explored as a surrogate of therapeutic and prognostic value.12-14 Specific inhibitor or vaccine targeting IDH mutations has also been reported to decrease 2-HG levels and release differentiation block and are evaluated as potential therapeutic strategy.15-17 Knock-in mice carrying IDH1-R132H mutation has been shown to increase numbers of early hematopoietic progenitors.18 However, the role of endogenous IDH1/2 on hematopoiesis, particularly during embryonic development, and whether the leukemogenic mechanisms of IDH1/2 mutations are conserved in other animal models are currently unclear.
Zebrafish has emerged as a model organism for the study of embryonic development and human diseases.19-21 During development, hematopoiesis occurs in 2 successive waves known as primitive and definitive hematopoiesis. Primitive erythropoiesis arises from the posterior lateral plate mesoderm at 12 hours postfertilization (hpf) that subsequently becomes the intermediate cell mass (ICM). Primitive myelopoiesis is first characterized by pu.1 expression at 12 hpf in the anterior lateral plate mesoderm, where it drives differentiation of hematopoietic cells toward the myeloid fate.22,23 Thereafter, the pu.1+ myeloid progenitor cells migrate to the yolk sac where they switch on expression of pan-leukocyte gene l-plastin as well as genes associated with macrophage (mpeg1, csf1ra, mfap4, cxcr3.2, and ptpn6)24 and neutrophil lineages (mpo and lyc).25,26 Definitive hematopoiesis arises from the ventral wall of the dorsal aorta (DA) at 36 hpf, as evident by c-myb and runx-1 expression.27-29 The definitive hematopoietic stem cell (HSC) then migrates to the caudal hematopoietic tissue (CHT), and then the kidney marrow, where life-long hematopoiesis occurs.30
In this study, we demonstrated for the first time the role of zidh1 during normal embryonic development and the conserved leukemogenic potential of human IDH1-R132H mutation in zebrafish embryos. The observations underscored the fundamental significance of normal and mutant IDH in the regulation of hematopoiesis and leukemia initiation. The availability of robust zebrafish model of human IDH1 mutation also opens up a new opportunity for a high-throughput drug-screening platform based on a whole-organism model.
Materials and methods
Zebrafish husbandry, embryos collection, and chemical treatment
Adult wild-type zebrafish and transgenic lines Tg(mpeg1:gfp), Tg(mpo:gfp), and Tg(flk1:gfp) were maintained under standard laboratory conditions at 28°C (Faculty Zebrafish Core Facility, Li Ka Shing Faculty of Medicine, the University of Hong Kong). Embryos were collected and staged according to Kimmel et al.31 1-Phenyl-2-thiourea (0.003%, Sigma) was used to suppress melanin formation after gastrulation. Zebrafish embryos were treated with human IDH1-R132H inhibitor, AGI-5198 (Xcess Biosciences) (10 μM), from bud stage (6 hpf) to 24 hpf. The study was approved by the Committee of the Use of Laboratory and Research Animals at the University of Hong Kong.
Cloning of zidh1, riboprobe synthesis, and WISH
Total RNA from 24-hpf embryos was extracted using TRIzol (Invitrogen) and reverse transcribed by the SuperScript II RT kit (Invitrogen). Primers used are shown in supplemental Table 1 on the Blood Web site. The zidh amplification products were cloned into pGEM-T Easy Vector (Promega) and used as template to generate antisense or sense (Shi and Leung, unpublished) digoxigenin-labeled RNA probes. Whole-mount in situ hybridization (WISH) of zebrafish embryos were performed as previously described32,33 using probes for zidh1, zidh2, scl, lmo2, pu.1, l-plastin, mpo, mpeg1, gata1, gata2, αhe1, cebpα, c-myb, rag1, efnb2α, and flt4. Probe for runx1 was a generous gift from Dr David Traver (University of California, San Diego).
Morpholinos, mRNA synthesis, and microinjection
Morpholinos (MOs, Gene-Tools, LLC) were designed to target the ATG codon of the zidh1 and zidh2 genes (zidh1MO and zidh2MO). Standard MO was used as negative controls. For messenger RNA (mRNA) rescue experiments, 5-bp mismatch primers were designed based on the MOs target-binding sequences without any amino acids change (supplemental Table 1). The polymerase chain reaction (PCR) amplicon was cloned into pGEM-T Easy Vector (Promega) and used as template to generate capped zidh1, zidh2 mRNAs with T7 or Sp6 RNA polymerase (mMessage mMachine kit, Ambion). For zebrafish embryo injections, MOs (6 ng) and capped mRNA (100 pg per embryo) were injected into 1- to 2-cell stage embryos at the yolk/cytoplasm boundary.
Construction of MO testing plasmid
A 1368-bp fragment of zidh1 transcript containing the MO target binding sequence was PCR amplified from complementary DNA of 24-hpf embryos (supplemental Table 1). The PCR product was purified with PCR purification kit (Qiagen) and cloned into the HindIII-BamHI sites of pegfp-N3 vector in-frame. Construct containing the 5′ untranslated region zidh1-gfp fusion cassette was bidirectionally sequenced and linearized for injecting into 1-cell stage embryos with or without zidh1MO.
qRT-PCR
Total RNA extraction and complementary DNA synthesis of zebrafish embryos were performed as described previously.34 Quantitative reverse-transcription PCR (qRT-PCR) was performed using the StepOnePlus Real-time PCR system (Applied Biosystems Inc.) with a SYBR Green master mix. Analysis was performed in triplicate by the 2−ΔΔC(T) method and β-actin was used as the housekeeping gene.35 A least 30 embryos were included in each experiment. The primer sequences are described in supplemental Table 1.
Transverse sectioning
The zebrafish embryos were collected and fixed overnight at 4°C in Bouin fixative. Ten-micrometer serial sections of paraffin-embedded fish were stained with hematoxylin and eosin as described.36
Flow cytometry and mpo+ cell sorting
Dissociation of embryos was performed as previously described.33 Briefly, dechorionated wild-type or transgenic embryos at the desired stage were collected and rinsed in E3 solution, digested with 0.05% trypsin-EDTA solution (Invitrogen) for 15 minutes at 28°C, and then completely dissociated to single-cell suspension by pipetting. CaCl2 (2 mM) was added to terminate the digestion. The whole cells were filtered through a 40-μm cell strainer (BD Falcon), spun down at 1200 rpm for 5 minutes at room temperature, then washed 2 times and harvested in phosphate-buffered saline with 2% (vol/vol) fetal bovine solution. The obtained cell suspension was subjected to Cytomics FC 500 (Beckman Coulter) at room temperature and enumerated by the percentage of GFP+ cells or sorted (MoFloTM XDP, Beckman Coulter) for examining their morphology with Wright-Giemsa staining.
Western blot, immunohistochemistry staining, whole-mount terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick end labeling staining, 5-bromo-2′-deoxyuridine incorporation assay, kidney marrow (KM) examination, and enzyme-linked immunosorbent assay for 5-hydroxymethylcytosine
The detailed protocols for these assays are described in the supplemental information.
Transcription activator–like effector nuclease (TALEN) design, construction, and somatic mutation analysis
Detailed protocols are described in the supplemental information.
Site-directed mutagenesis assay
Total RNA from human embryonic kidney 293T cells were extracted by using TRIzol (Invitrogen) and reverse transcribed by the SuperScript II RT kit (Invitrogen) following the manufacturer’s recommendations. Based on published sequence of IDH1 in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/), a pair of primers (supplemental Table 1) was designed to amplify the coding region by using Platinum Pfx DNA polymerase (Invitrogen). The PCR product was cloned downstream of ubiquitously expressed cytomegalovirus promoter in the pCS2+ vector34,37 and bidirectionally sequenced. The zebrafish idh1 was ligated in-frame at its C terminus with egfp that was derived from pegfp-N3 vector. The construct of zidh1-WT-egfp was cloned into the ClaI and EcoRI sites of pCS2+ vector and bidirectionally sequenced. Site-directed mutagenesis was performed using QuikChange Site-Directed Mutagenesis Kit (Stratagene) to introduce IDH1-R132H and zidh1-R146H mutations and confirmed by direct sequencing. Primers used are shown in supplemental Table 1. All the construction were linearized and injected into 1-cell stage zebrafish embryos.
Metabolite extraction and GC-MS
The determination of 2-HG was conducted as described previously.5,38 Metabolite extraction was performed by adding ice-cold 80% methanol (with 20% water) to embryos at 24 hpf followed by sonication (3 seconds on and 3 seconds off for 5 minutes) on ice using the Ultrasonic Processor (Cole-Parmer). Methanol extracts were incubated at −80°C for 30 minutes and centrifuged at top speed for 30 minutes at 4°C to remove precipitated protein and cellular debris. The aqueous metabolites in the supernatant layer were evaporated to dryness under nitrogen gas and stored at −80°C.
For gas chromatography-mass spectrometry (GC-MS) analysis, dried extracts were resuspended in equal volume mixture of acetonitrile and N-methyl-N-tert-butyldimethylsilyltrifluoroacetamide (MTBSTFA, Regis) and heated at 70°C for 75 minutes. An aliquot of derivatized materials (1 μL) was injected into a 450-GC (Bruker) equipped with a VF-5 ms capillary column (30 m × 0.25 mM, inner wall of small diameter × 0.25 μM film thickness, Varian, Netherlands), connected to a 320-MS triple quadrupole (Bruker) in a splitless model. The injection temperature, interface temperature, and the ion source temperature were set to 310°C, 250°C, and 200°C, respectively. The initial GC oven temperature was set at 100°C for 3 minutes, and was raised to 230°C at 4°C/minute and held for 4 minutes, then ramped up to 300°C and maintained for 5 minutes. The measurements were carried out with electronic ionization (70 eV) in the full-scan mode (m/z: 50-550 amu) and detector voltage was set to 1200 V. Identification of the 2-HG peak was confirmed by using standards purchased from Sigma-Aldrich. The 2-HG and glutamate signal intensities were quantified by integration of peak areas. Relative quantification of 2-HG was determined by the ratio of signal intensities for 2-HG normalized to glutamate and corrected by embryo numbers.
Imaging analysis
Embryos were examined using a Nikon SMZ800 stereomicroscope (Nikon Hong Kong Ltd.) and photographed by a Nikon Digital Sight DS-Fi1 camera (Nikon Hong Kong Ltd.). Images of blood smear samples were acquired on an Olympus IX70 microscope (Olympus). All the images were processed with Adobe Photoshop, version 7.0 (Adobe, San Jose).
Statistical analysis
The data were expressed as mean ± standard error of the mean and were analyzed by the Student t test. A P value of less than .05 was considered statistically significant.
Results
IDH1 is highly conserved
IDH1 is homodimeric NAD phosphate+–dependent enzyme that catalyzes decarboxylation of isocitrate to produce α-ketoglutarate, reduced NAD phosphate, and CO2. Mammalian and zebrafish IDH1 proteins are evolutionarily conserved, including an isocitrate and isopropylmalate dehydrogenases signature sequence with glycine-rich residues, 7 amino acid residues involved in isocitrate-Mg2+ binding, and a C-terminal peroxisomal-targeting signal39 (supplemental Figure 1A). The amino acid sequence of zidh1 protein shares an overall 83% and 84% identity with its human and mouse orthologs. Phylogenetic analysis shows that zidh1 clustered closely with mouse and human IDH1 and separated from groups of IDH2 or IDH3 protein sequences (supplemental Figure 1B). Furthermore, zidh1 and zidh2 are located on chromosomes 9 and 18, and their syntenic neighboring genes are conserved in the locus of human IDH1 and IDH2 on chromosomes 2 and 15 (Figure 1A). These results indicate that zidh1/2 are true orthologs of mammalian IDH1/2.
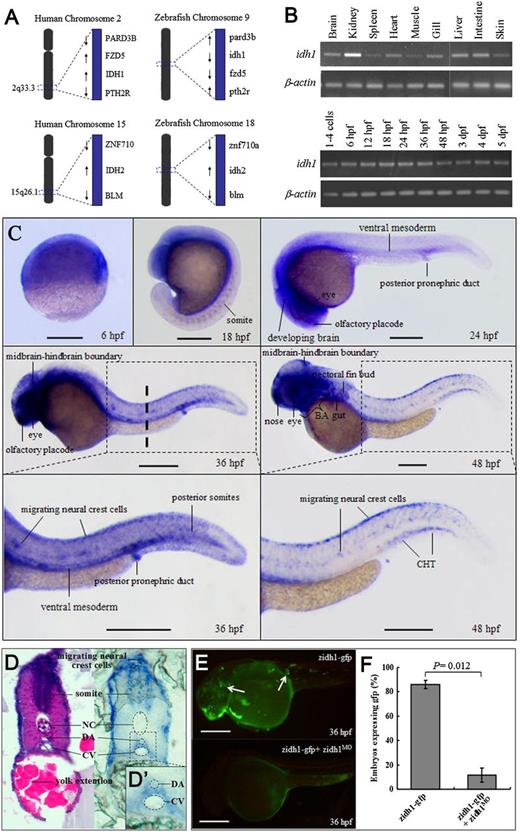
Spatiotemporal expression of zidh1 during zebrafish development. (A) The genomic loci surrounding human IDH1/2 in chromosomes 2q33 and 15q26.1 are syntenic with the regions of zidh1/2 in chromosomes 9 and 18. (B) Semi-qRT-PCR of zidh1 in different adult tissues (brain, kidney, spleen, heart, muscle, gill, liver, intestine, and skin) and at different embryonic stages normalized to β-actin. (C) Spatial expression of zidh1. Representative figures from at least 3 separate experiments containing more than 10 embryos per experiment are shown. Lateral view, anterior to the left, dorsal upwards. Dashed line indicates the level of the histological section in panel D. (D) Paraffin section at 36 hpf showing zidh1 expression around the ventral wall of the DA region (inset D′). (E) Mosaic fluorescent pattern (arrows) in embryos injected with linearized zidh1-gfp plasmid at 1-cell stage are almost completely abolished (lower panel) by zidh1MO coinjection. (F) The percentages of embryos expressing gfp with or without zidh1MO coinjection are enumerated. NC, notochord; CV, caudal vein. BA, brachial arches; CHT, caudal hematopoietic tissue. These data were performed in 3 independent experiments. Scale bars represent 250 μm.
Spatiotemporal expression of zidh1 during zebrafish development. (A) The genomic loci surrounding human IDH1/2 in chromosomes 2q33 and 15q26.1 are syntenic with the regions of zidh1/2 in chromosomes 9 and 18. (B) Semi-qRT-PCR of zidh1 in different adult tissues (brain, kidney, spleen, heart, muscle, gill, liver, intestine, and skin) and at different embryonic stages normalized to β-actin. (C) Spatial expression of zidh1. Representative figures from at least 3 separate experiments containing more than 10 embryos per experiment are shown. Lateral view, anterior to the left, dorsal upwards. Dashed line indicates the level of the histological section in panel D. (D) Paraffin section at 36 hpf showing zidh1 expression around the ventral wall of the DA region (inset D′). (E) Mosaic fluorescent pattern (arrows) in embryos injected with linearized zidh1-gfp plasmid at 1-cell stage are almost completely abolished (lower panel) by zidh1MO coinjection. (F) The percentages of embryos expressing gfp with or without zidh1MO coinjection are enumerated. NC, notochord; CV, caudal vein. BA, brachial arches; CHT, caudal hematopoietic tissue. These data were performed in 3 independent experiments. Scale bars represent 250 μm.
Expression pattern of zidh1 gene
In adult, zidh1 was highly expressed in kidney among other tissues (Figure 1B). In embryos, zidh1 was expressed as early as the 1-cell stage, indicative of the presence of maternal transcript, and throughout the first 5 days of development (Figure 1B). At 6 hpf, zidh1 was ubiquitously expressed and at 18 to 24 hpf, it became more restricted to the developing brain, eye, somites, olfactory placode, ventral mesoderm, and posterior pronephric duct (Figure 1C). At 36 hpf, zidh1 was also expressed in the migrating neural crest cells and around the ventral wall of DA (Figure 1D-D′). At 48 hpf, zidh1 was more specifically expressed in the nose, eye, midbrain-hindbrain boundary, brachial arches, gut, pectoral fin bud, migrating neural crest cells as well as the hematopoietic site CHT region (Figure 1C).
Knockdown of zidh1 gene disrupts primitive myelopoiesis
To examine the role of zidh1 during embryonic hematopoiesis, the zidh1 gene was knockdown by morpholino targeting its ATG codon (zidh1MO). More than 90% of the embryos could tolerate up to 6 ng without obvious toxicity and lethality, and this dose was used in all subsequent experiments, although a small portion of embryos exhibited reduced head, body-axis curvature, and reduced yolk extension at 48 hpf (supplemental Figure 2A-B). The latter were excluded from subsequent analysis. To ascertain the effectiveness of molecular targeting, we injected a zidh1-gfp chimeric plasmid containing a zidh1MO-targeted sequence at the 5′ terminus into 1-cell stage embryos and demonstrated mosaic gfp expression pattern, which was significantly quenched by coinjecting zidh1MO (percentage of embryos expressed egfp in zidh1-gfp, 85.9% ± 3.4%; in zidh1-gfp + zidh1MO, 11.58% ± 5.9%, P = .012) (Figure 1E-F).
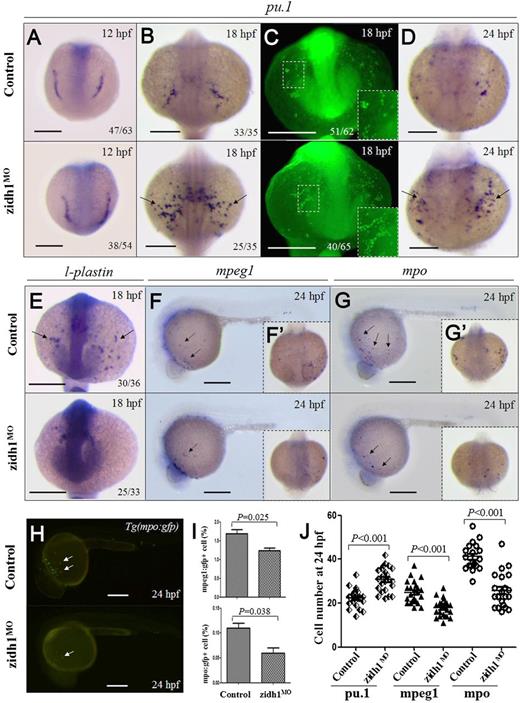
The effects of zidh1 knockdown on myelopoiesis were examined. At 12 hpf, the expression of pu.1 was unchanged in the zidh1MO embryos (Figure 2A). However, at 18 to 24 hpf, pu.1 expression was significantly increased (Figure 2B-D). The results were confirmed by qRT-PCR (supplemental Table 2) and immunostaining (Figure 2C). On the other hand, genes associated with differentiated myeloid cells (l-plastin, mpo, and mpeg1) were significantly reduced as shown by WISH (Figure 2E-G) as well as direct inspection and flow cytometric analysis of dissociated Tg(mpo:gfp) and Tg(mpeg1:gfp) embryos (Figure 2H-I; supplemental Figure 3). The effects of zidh1 knockdown on primitive myelopoiesis could be rescued by wild-type zidh1 but not the zebrafish ortholog of human IDH1R132H (zidh1-R146H) mRNA (supplemental Table 2; supplemental Figure 4). Similar changes in myeloid gene expression were seen in the CHT at 36 hpf and 3 days postfertilization (dpf) (supplemental Figure 5). No increase in apoptotic signal was detectable in anterior lateral plate mesoderm or posterior ICM (pICM) (supplemental Figure 6). Expression of genes associated with early hematopoietic precursor (scl and lmo2), erythropoiesis (gata1, α-he1, and β-he1) (supplemental Figure 2D) and vascular endothelial cells (fli1) was not affected (supplemental Table 2). Expression of c/ebpα and gfi-1 was unaffected in the zidh1MO embryos (supplemental Table 2).
zidh1 knockdown perturbed myeloid development. Expression of pu.1 mRNA (A,B,D) and protein (C) in control and zidh1MO embryos at 12, 18, or 24 hpf. (C) Higher magnification of the yolk sac is shown in the insets. The numbers of embryos with the typical pu.1 immunostaining out of the total number of embryos evaluated are shown. mRNA expression of l-plastin (E), mpeg1 (F,F′), and mpo (G,G′) at 18 or 24 hpf. (H) Fluorescent microscopy of Tg(mpo:gfp) at 24 hpf showing decreased gfp+ cells after zidh1 knockdown. (I) Percentage of mpeg1:gfp+ and mpo:gfp+ cells in dissociated control and zidh1MO embryos at 24 hpf by flow cytometry (3 independent experiments, representative results are shown in supplemental Figure 3). (J) The cell numbers of pu.1, mpeg1, and mpo positive cells at 24 hpf were counted; each symbol represents the cell number in 1 embryo. Scale bars represent 250 μm.
zidh1 knockdown perturbed myeloid development. Expression of pu.1 mRNA (A,B,D) and protein (C) in control and zidh1MO embryos at 12, 18, or 24 hpf. (C) Higher magnification of the yolk sac is shown in the insets. The numbers of embryos with the typical pu.1 immunostaining out of the total number of embryos evaluated are shown. mRNA expression of l-plastin (E), mpeg1 (F,F′), and mpo (G,G′) at 18 or 24 hpf. (H) Fluorescent microscopy of Tg(mpo:gfp) at 24 hpf showing decreased gfp+ cells after zidh1 knockdown. (I) Percentage of mpeg1:gfp+ and mpo:gfp+ cells in dissociated control and zidh1MO embryos at 24 hpf by flow cytometry (3 independent experiments, representative results are shown in supplemental Figure 3). (J) The cell numbers of pu.1, mpeg1, and mpo positive cells at 24 hpf were counted; each symbol represents the cell number in 1 embryo. Scale bars represent 250 μm.
Knockdown of zidh1 gene reduces definitive hematopoiesis
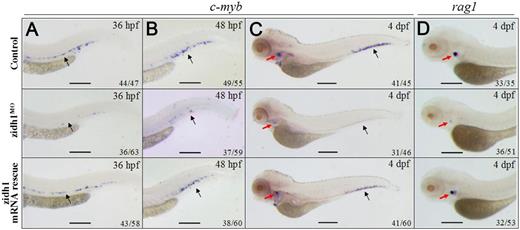
Knockdown of zidh1 resulted in significant reduction of runx-1 (supplemental Figure 7) and c-myb expression in the ventral wall of DA at 36 and 48 hpf (Figure 3A-B) and at the CHT region at 4 dpf (Figure 3C-D). The results were confirmed by qRT-PCR (supplemental Table 3). Expression of T-cell marker rag1 in the thymus was also reduced (Figure 3D). Apoptosis was not increased at the vicinity of c-myb expression at 36 or 48 hpf (supplemental Figure 8). Coinjecting a wild-type zidh1 mRNA resistant to MO into 1-cell stage embryos reversed the phenotypes of the zidh1MO embryos, including the upregulation of pu.1 and downregulation of c-myb (supplemental Table 3) and rag1 expression (Figure 3A-D), confirming the specificity of MO targeting. Gene expression associated with arterial and venous specification as well as the axial circulation was unaffected in zidh1MO embryos (supplemental Figure 9).
zidh1 knockdown perturbed definitive hematopoiesis. Expression of c-myb in control (upper panel), zidh1MO embryos with (lower panel), or without (middle panel) zidh1 mRNA rescue at the ventral wall of the DA (black arrows) at 36 hpf (A) and at the caudal hematopoietic tissues (black arrows) at 48 hpf (B) and 4 dpf (C). The red arrows in panel C indicate c-myb expression in the thymus. (D) Expression of rag1 (red arrows) in control (upper panel), zidh1MO embryos with (lower panel), or without (middle panel) zidh1 mRNA rescue at 4 dpf. These data were performed in 3 independent experiments. Scale bars represent 250 μm.
zidh1 knockdown perturbed definitive hematopoiesis. Expression of c-myb in control (upper panel), zidh1MO embryos with (lower panel), or without (middle panel) zidh1 mRNA rescue at the ventral wall of the DA (black arrows) at 36 hpf (A) and at the caudal hematopoietic tissues (black arrows) at 48 hpf (B) and 4 dpf (C). The red arrows in panel C indicate c-myb expression in the thymus. (D) Expression of rag1 (red arrows) in control (upper panel), zidh1MO embryos with (lower panel), or without (middle panel) zidh1 mRNA rescue at 4 dpf. These data were performed in 3 independent experiments. Scale bars represent 250 μm.
zidh1 TALENs recapitulate hematopoietic function of zidh1MO
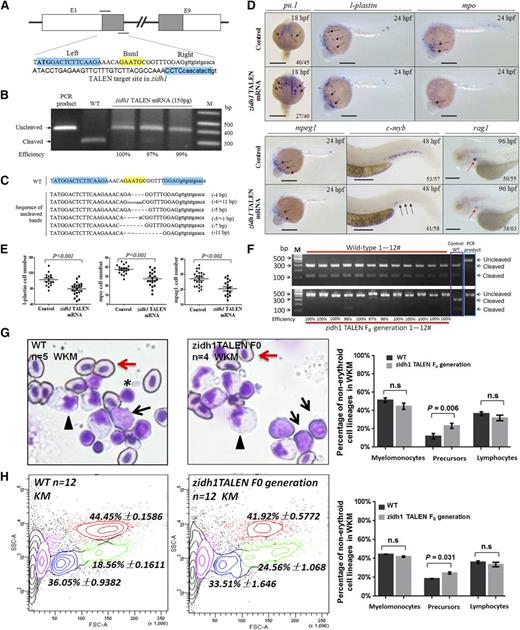
We further constructed a TALEN pair targeting exon 1 of zidh1 with a BsmI restriction site for restriction fragment length polymorphism assay (Figure 4A). Transcribed mRNAs encoding the zidh1 TALEN pair were injected into the cytoplasm of wild-type zebrafish embryos at the 1-cell stage. At 150 pg (each arm), 85% to 90% of the injected embryos developed normally. To evaluate the mutagenic efficiency, a 410-bp genomic DNA sequence containing the target site from 24-hpf embryos was amplified and digested with BsmI. Embryos injected with zidh1 TALEN mRNAs exhibited a loss of BsmI site with an efficiency of 97% (Figure 4B), resulting from small indels of various lengths at targeting locus (Figure 4C). These embryos showed a significant increase in pu.1 but a decrease in l-plastin, mpo, and mpeg1 expression (Figure 4D-E). Definitive hematopoiesis was reduced as evident by the downregulation of c-myb and rag1 expression (Figure 4D). The expression of scl, lmo2, gata1, and α-he1 was unchanged (supplemental Figure 10). The results recapitulated the hematopoietic phenotypes seen in the zidh1MO embryos, providing strong evidence that zidh1 is important for myeloid differentiation and maintenance of definitive HSC. The F0 larvae were raised to adulthood (3 months) and genotype confirmed nearly 100% knockout efficiency in all 12 fish examined (Figure 4F). KM was examined in these and wild-type adult fish, showing a significant increase in precursor cells, characterized by relatively large size with high nuclear/cytoplasmic ratio, in the zidh1 TALEN fish (Figure 4G). There was no change in myelomonocytic or lymphoid populations. The results were confirmed by flow cytometry (Figure 4H).
Targeted mutation of zidh1 using TALENs recapitulated the hematopoietic defects of zidh1MO. (A) Target site of zidh1 by TALEN. The binding sequence for the TALEN pair were highlighted in cyan (indicated by “left” and “right”) and the BsmI site in yellow. The start codon of zidh1 was bold. E, exon. (B) BsmI digestion of PCR products from pooled genomic DNA of 5 embryos injected with zidh1 TALEN mRNAs in 3 experiments. The cleaved and uncleaved PCR products are indicated. M, marker; WT, wild-type. (C) Representative sequences of uncleaved PCR products. (D) Expression of pu.1 at 18 hpf, l-plastin, mpo, and mpeg1 at 24 hpf; c-myb at 48 hpf; and rag1 at 96 hpf in control and zidh1 TALEN mRNA-injected embryos. (E) The numbers of l-plastin–, mpo-, and mpeg1-positive cells per embryos were counted and are expressed as mean ± standard error of the mean. The numbers in each panel indicates the number of embryos with representative phenotype out of the total number of embryos evaluated. Bars represent 250 μm. (F) Genotyping of 12 zidh1 TALEN and wild-type fish at 3 months old, showing near 100% knockout efficiency based on Bsml digestion of PCR products. The uncleaved (TALENed) and cleaved PCR products are shown. (G, left and middle) May-Grunwald/Giemsa staining of the KM showing hematopoietic cells of different morphologic lineage. Arrowheads, myelomonocytes; asterisks, lymphocytes; black arrows, precursor cells; red arrows, erythrocytes. Original magnification, ×400. (G, right) Mean percentage of nonerythroid cells in the KM, showing statistically significant increase in precursor cell population (5 wild-type and 4 zidh1 TALEN F0 fish were examined). (H, left and middle) Contour plot showing the myelomonocytic (red), precursor (green), lymphocytic (blue), and erythroid lineages (purple). (H, right) The histogram shows the average results of 12 wild-type and TALEN F0 adult fish. n.s., not significant.
Targeted mutation of zidh1 using TALENs recapitulated the hematopoietic defects of zidh1MO. (A) Target site of zidh1 by TALEN. The binding sequence for the TALEN pair were highlighted in cyan (indicated by “left” and “right”) and the BsmI site in yellow. The start codon of zidh1 was bold. E, exon. (B) BsmI digestion of PCR products from pooled genomic DNA of 5 embryos injected with zidh1 TALEN mRNAs in 3 experiments. The cleaved and uncleaved PCR products are indicated. M, marker; WT, wild-type. (C) Representative sequences of uncleaved PCR products. (D) Expression of pu.1 at 18 hpf, l-plastin, mpo, and mpeg1 at 24 hpf; c-myb at 48 hpf; and rag1 at 96 hpf in control and zidh1 TALEN mRNA-injected embryos. (E) The numbers of l-plastin–, mpo-, and mpeg1-positive cells per embryos were counted and are expressed as mean ± standard error of the mean. The numbers in each panel indicates the number of embryos with representative phenotype out of the total number of embryos evaluated. Bars represent 250 μm. (F) Genotyping of 12 zidh1 TALEN and wild-type fish at 3 months old, showing near 100% knockout efficiency based on Bsml digestion of PCR products. The uncleaved (TALENed) and cleaved PCR products are shown. (G, left and middle) May-Grunwald/Giemsa staining of the KM showing hematopoietic cells of different morphologic lineage. Arrowheads, myelomonocytes; asterisks, lymphocytes; black arrows, precursor cells; red arrows, erythrocytes. Original magnification, ×400. (G, right) Mean percentage of nonerythroid cells in the KM, showing statistically significant increase in precursor cell population (5 wild-type and 4 zidh1 TALEN F0 fish were examined). (H, left and middle) Contour plot showing the myelomonocytic (red), precursor (green), lymphocytic (blue), and erythroid lineages (purple). (H, right) The histogram shows the average results of 12 wild-type and TALEN F0 adult fish. n.s., not significant.
Hematopoietic function redundancy for zidh1 and zidh2
The mutual exclusion of IDH1/2 mutations in AML suggested that the 2 genes may share the same pathogenetic pathway.7 Using the zebrafish model, functional redundancy between zidh1 and zidh2 during embryogenesis was evaluated. Unlike zidh1, zidh2 was first expressed in the developing head and eye at 18 hpf and more ubiquitously expressed in the whole embryos at 24 hpf. It was expressed preferentially in the eye, developing brain, brachial arch, posterior pronephric duct, and intersomitic region at 36 hpf. Thereafter, zidh2 expression became more restricted to the pectoral fin bud and brain at 48 hpf (supplemental Figure 11A). zidh2 knockdown resulted in significant, albeit modest, reduction in gene expression associated with primitive erythropoiesis (supplemental Table 4; supplemental Figure 11B). Importantly, genes associated with early myeloid progenitor (pu.1) development were also increased, whereas those with late myeloid (l-plastin, mpo) development were decreased reminiscent of the changes induced by zidh1 knockdown. Definitive hematopoiesis was not affected (supplemental Figure 11C). The increase in pu.1 expression in zidh1MO embryos was only partially rescued by zidh2 mRNA, whereas the diminished definitive hematopoiesis was unaffected (supplemental Table 3). These observations supported the proposition that zidh2 might play a role in the regulation of embryonic hematopoiesis distinctive from zidh1 during zebrafish development.
Overexpression of human IDH1-R132H causes expanded embryonic hematopoiesis
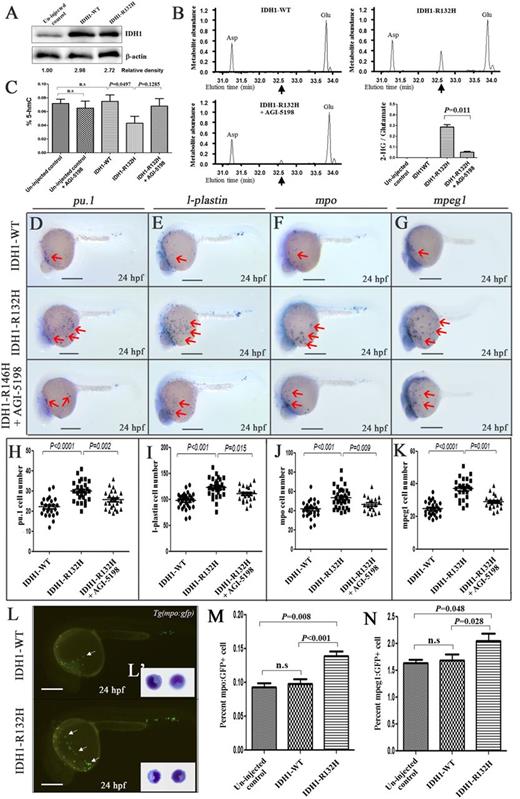
To ascertain the fundamental role in human IDH1-R132H in leukemia initiation, plasmids of IDH1-R132H and IDH1-WT were injected into 1-cell stage embryos at 100 pg. The 2 groups of embryos were morphologically indistinguishable at 24 hpf after injection. Western blotting and GC-MS analyses on pooled embryos at 24 hpf confirmed strong protein expression of IDH1 (Figure 5A) and elevated 2-HG levels in IDH1-R132H injected embryos (Figure 5B). 5-Hydroxymethylcytotsine (5-hmC), converted from 5-methylcytosine by ten eleven translocation 2 (TET2), was reduced (Figure 5C). Importantly, the effects of IDH1-R132H expression on 2-HG and 5-hmC could be reversed by AGI-5198, a specific inhibitor of IDH1-R132H that had no effect on normal hematopoiesis (supplemental Figure 12). The hematopoietic phenotype of IDH1-R132H expression was examined. At 24 hpf, approximately 25% IDH1-R132H–injected embryos exhibited an expansion of the pICM. That was not found in embryos injected with IDH1-WT (supplemental Figure 13A-D). The pICM was positive for gata1 expression and 5-bromo-2′-deoxyuridine incorporation and could be inhibited by AGI-5198. Furthermore, IDH1-R132H–injected embryos showed increased pu.1, l-plastin, mpo and mpeg1 expression in the yolk sac at 24 hpf as shown by WISH (Figure 5D-K) as well as flow cytometry of dissociated Tg(mpo:gfp) and Tg(mpeg1:gfp) transgenic lines (Figure 5L-N; supplemental Figure 14). The mpo-gfp+ cells isolated from IDH1-R132H–injected embryos had the morphological features of primitive myeloid cells (Figure 5L′). A similar increase in pu.1, l-plastin, mpo, and c-myb expression was seen at 36 hpf (supplemental Figure 15A-G).
Effects of human IDH1-R132H overexpression. (A) IDH1 protein expression in uninjected and IDH1-WT– or IDH1-R132H–injected embryos at 24 hpf. The numbers indicate the relative band intensity as quantified by Image J software and normalized to levels of β-actin. (B) Representative gas chromatographs from IDH1-WT– or IDH1-R132H–injected embryos with or without AGI-5198 treatment. The spikes represent the MTBSTFA-derivatized intracellular metabolites eluting between 31.0 and 34.0 minutes, including aspartate (Asp), 2-HG, and glutamate (Glu). Black arrows indicate the expected elution time of 32.6 minutes for MTBSTFA-derivatized 2-HG. Metabolite abundance refers to GC-MS signal intensity. Quantitation of 2-HG signal intensities relative to the intrasample glutamate signals of 3 different experiments was shown. (C) 5-hydroxymethylcytotsine (5-hmC) was reduced upon IDH-R132H expression. Expression of early myeloid progenitor marker pu.1 (D), and late myelomonocytic differentiation markers l-plastin (E), mpo (F), and mpeg1 (G) at 24 hpf in embryos injected with complementary DNA encoding IDH1-WT, or IDH1-R132H, with or without AGI-5198 (10 μM) treatment starting from bud stage. The red arrows indicate myeloid cells on the yolk sac. (H-K) The cell numbers of pu.1 (H), l-plastin– (I), mpo- (J), and mpeg1- (K) positive cells per embryos. (L) Fluorescent microscopy of Tg(mpo:gfp) embryos at 24 hpf showing increased gfp+ cells in embryos injected with IDH1-R132H compared with IDH1-WT. The inset (L′) showed the morphology of gfp+ cells in dissociated Tg(mpo:gfp) embryos at 36 hpf. (M,N) Percentage of mpeg1:GFP+ and mpo:gfp+ cells in uninjected control–, IDH1-WT–, and IDH1-R132H–injected embryos at 24 hpf as enumerated by flow cytometry (representative results are shown in supplemental Figure 11) in 3 different experiments. n.s., not significant. Bars represent 250 μm and ×600 magnification in panel (L′).
Effects of human IDH1-R132H overexpression. (A) IDH1 protein expression in uninjected and IDH1-WT– or IDH1-R132H–injected embryos at 24 hpf. The numbers indicate the relative band intensity as quantified by Image J software and normalized to levels of β-actin. (B) Representative gas chromatographs from IDH1-WT– or IDH1-R132H–injected embryos with or without AGI-5198 treatment. The spikes represent the MTBSTFA-derivatized intracellular metabolites eluting between 31.0 and 34.0 minutes, including aspartate (Asp), 2-HG, and glutamate (Glu). Black arrows indicate the expected elution time of 32.6 minutes for MTBSTFA-derivatized 2-HG. Metabolite abundance refers to GC-MS signal intensity. Quantitation of 2-HG signal intensities relative to the intrasample glutamate signals of 3 different experiments was shown. (C) 5-hydroxymethylcytotsine (5-hmC) was reduced upon IDH-R132H expression. Expression of early myeloid progenitor marker pu.1 (D), and late myelomonocytic differentiation markers l-plastin (E), mpo (F), and mpeg1 (G) at 24 hpf in embryos injected with complementary DNA encoding IDH1-WT, or IDH1-R132H, with or without AGI-5198 (10 μM) treatment starting from bud stage. The red arrows indicate myeloid cells on the yolk sac. (H-K) The cell numbers of pu.1 (H), l-plastin– (I), mpo- (J), and mpeg1- (K) positive cells per embryos. (L) Fluorescent microscopy of Tg(mpo:gfp) embryos at 24 hpf showing increased gfp+ cells in embryos injected with IDH1-R132H compared with IDH1-WT. The inset (L′) showed the morphology of gfp+ cells in dissociated Tg(mpo:gfp) embryos at 36 hpf. (M,N) Percentage of mpeg1:GFP+ and mpo:gfp+ cells in uninjected control–, IDH1-WT–, and IDH1-R132H–injected embryos at 24 hpf as enumerated by flow cytometry (representative results are shown in supplemental Figure 11) in 3 different experiments. n.s., not significant. Bars represent 250 μm and ×600 magnification in panel (L′).
Overexpression of zidh1-R146H mutation expands myelopoiesis
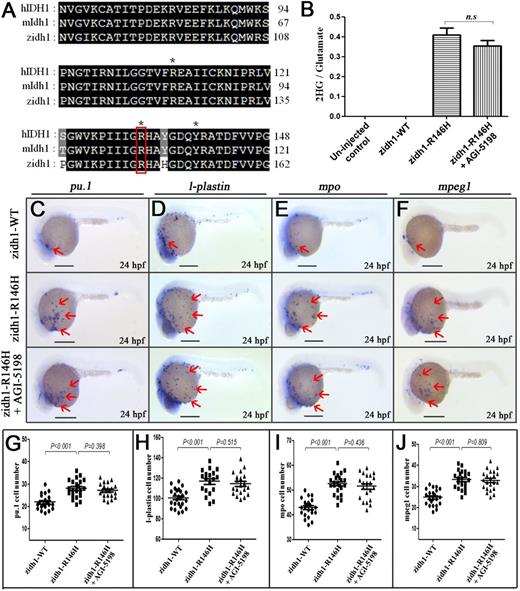
To demonstrate the specific pathogenetic role of R to H substitution in IDH1-R132H, we performed site-directed mutagenesis and generated a zidh1-R146H mutant corresponding to the human IDH1-R132H mutation (Figure 6A). Injection of zidh1-WT-egfp and zidh1-R146H-egfp resulted in mosaic egfp expression pattern, as shown by the presence of egfp+ cells in the anterior mesoderm and pICM region (supplemental Figure 16). Overexpression of zidh1-R146H but not wild-type zidh1 significantly increased 2-HG level in zebrafish embryos (Figure 6B) and resulted in myeloid expansion, characterized by the expression of pu.1, l-plastin, mpo, and mpeg1 (Figure 6C-J). The myeloid expansion and rise in 2-HG induced by zidh1-R146H was not affected by AGI-5198 treatment.
Overexpression of zidh1-R146H mutation-induced expansion of myeloid compartments. (A) Amino acid sequence alignment of IDH1 protein among human (h), mouse (m), and zebrafish (z), containing the cancer-associated residue R, which are marked by red box. *Residues involved in isocitrate binding. (B) Quantitation of 2-HG signal intensities relative to the glutamate signals of the same samples from 3 experiments. n.s., not significant. (C-F) Expression of pu.1 (C), l-plastin (D), mpo (E), and mpeg1 (F) in zidh1-WT-egfp– and zidh1-R146H-egfp–injected embryos at 24 hpf; the latter was treated with AGI-5198 (10 μM). The red arrows indicate myeloid cells on the yolk sac. The numbers of pu.1 (G), l-plastin (H), mpo (I), and mpeg1 (J) positive cells per embryos were manually counted. Scale bars represent 250 μm.
Overexpression of zidh1-R146H mutation-induced expansion of myeloid compartments. (A) Amino acid sequence alignment of IDH1 protein among human (h), mouse (m), and zebrafish (z), containing the cancer-associated residue R, which are marked by red box. *Residues involved in isocitrate binding. (B) Quantitation of 2-HG signal intensities relative to the glutamate signals of the same samples from 3 experiments. n.s., not significant. (C-F) Expression of pu.1 (C), l-plastin (D), mpo (E), and mpeg1 (F) in zidh1-WT-egfp– and zidh1-R146H-egfp–injected embryos at 24 hpf; the latter was treated with AGI-5198 (10 μM). The red arrows indicate myeloid cells on the yolk sac. The numbers of pu.1 (G), l-plastin (H), mpo (I), and mpeg1 (J) positive cells per embryos were manually counted. Scale bars represent 250 μm.
Discussion
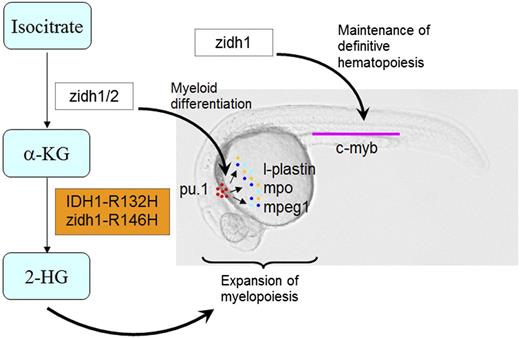
In this study, we took advantage of the zebrafish model to examine the hitherto undescribed role of idh1 in embryonic hematopoiesis and its relevance to human AML (Figure 7). With this model, the spatiotemporal expression of zidh1 during embryogenesis has been shown in detail, in particular at the vicinity of definitive HSC at ventral wall of DA. Gene perturbation by morpholino and TALEN consistently demonstrated that zidh1-modulated myeloid differentiation and was involved in the initiation and maintenance of definitive HSC. The leukemogenic pathways operative in IDH1-R132H+ AML were conserved in zebrafish. These observations have provided important insights to the mechanisms of embryonic hematopoiesis and leukemogenesis.
A diagrammatic presentation of the roles of zidh1/2 and IDH1-R132H mutation in primitive myelopoiesis and definitive hematopoiesis during zebrafish embryonic development. zidh1 and zidh2 played redundant roles in regulating myeloid differentiation, whereas zidh1 but not zidh2 also played a role in the maintenance of definitive hematopoiesis. Human IDH1-R132H and zebrafish idh1-R146H resulted in an increased 2-HG that induced expansion of primitive myelopoiesis.
A diagrammatic presentation of the roles of zidh1/2 and IDH1-R132H mutation in primitive myelopoiesis and definitive hematopoiesis during zebrafish embryonic development. zidh1 and zidh2 played redundant roles in regulating myeloid differentiation, whereas zidh1 but not zidh2 also played a role in the maintenance of definitive hematopoiesis. Human IDH1-R132H and zebrafish idh1-R146H resulted in an increased 2-HG that induced expansion of primitive myelopoiesis.
First, the present study demonstrated for the first time the role of zidh1 in modulating myeloid differentiation during embryonic hematopoiesis. In particular, loss of function of zidh1 by both morpholino knockdown and TALEN-mediated mutagenesis increased the number of cells with pu.1 expression (early myeloid progenitor) and decreased those with mpo and mpeg1 expression (granulocytes and macrophages). The results were specific and could be rescued by coinjection of wild-type zidh1 mRNA. However, the dynamics of changes in gene expression and its impact on cell fate decision on a cellular basis were not examined. There was no increase in apoptosis of the mpo and mpeg1 expressing cells, suggesting that zidh1 suppression/deletion resulted in blockade of differentiation of the myeloid lineage. In addition, there was an increase in mcsf expression in zidhMO embryos (supplemental Table 5), which has been shown to induce pu.1 expression in zebrafish embryos.40 An increase in precursor cells was shown in adult zidh1 TALEN F0 fish, associated with a modest decrease in terminally differentiated myelomonocytic and lymphoid components which was not statistically significant. The results corroborated with the increase in pu.1 and decrease in mpo and mpeg1 expression in zebrafish embryos and provided a possible mechanistic link between IDH1 mutation, loss of IDH1 function, and differentiation block in AML.
Second, we demonstrated for the first time the role of zidh1 in definitive hematopoiesis. In particular, zidh1 was expressed at the vicinity of ventral wall of DA, where definitive HSCs arise and both zidh1 knockdown and targeted mutagenesis previously mentioned significantly decreased c-myb expression in the ventral wall of DA and CHT as well as rag1 expression in the thymus. zidh1 knockdown had no effect on the expression of target genes downstream of Wnt, Notch, or transforming growth factor-β or the protein expression of β-catenin (Wnt), Notch1 intracellular domain (Notch), and phospho-Smad2 (transforming growth factor-β) (supplemental Table 6; supplemental Figure 17), supporting the proposition that zidh1 might have a direct effect on HSCs rather than an effect on the upstream regulatory signals. Whether the effects on HSCs were caused by a perturbation of intermediary metabolism after zidh1 suppression/deletion or through a yet-identified signaling pathway, would have to be further investigated. Intriguingly, knockdown of zidh1 did not affect scl and lmo2 expression in the ICM, suggesting a lack of effect on primitive hematopoietic precursors.
Third, the results of human IDH1-R132H expression demonstrated that the pathways operative in human leukemia were conserved in zebrafish embryos. Human AML carrying a IDH1/2 mutation had impaired TET2 activity and exhibited DNA hypermethylation.7 In this study, overexpression of the human clone led to an increase in 2-HG level, a reduction of 5-hmC, and expansion of myelopoiesis and the responses were blockable by AGI-5198, an inhibitor specific for human IDH1-R132H mutant enzyme.15 Site-direct mutagenesis of zebrafish idh1 similarly induced 2-HG increase and myeloid expansion, but both responses were unaffected by AGI-5198. The observations supported the proposition, albeit indirectly, that an increase in 2-HG induced the myeloid expansion in zebrafish embryos. Direct treatment of zebrafish embryos with 2-HG was ineffective, reflecting its impermeable nature across the cell membrane.8 In fact, increase in 2-HG per se, as in patients with 2-HG aciduria, a condition in which serum 2-HG is increased because of homozygous loss-of-function mutations of 2-HG dehydrogenase enzymes, has not been known to cause leukemia.11 The cooperative event leading to leukemogenesis is presently unclear and, in addition to the block in myeloid differentiation as a result of loss of function of zidh1, it is likely dependent on concomitant genetic aberration including internal tandem duplication of Fms-like tyrosine kinase 3 (FLT3). The complexity is highlighted by the different impact of IDH1-R132H mutations on clinical outcome between glioma and AML11,41-43 and the apparent inconsistent results in the latter. To this end, co-injection of human IDH1-R132H and FLT3-internal tandem duplication plasmids into zebrafish embryos, at low doses that did not induce myeloid expansion on their own, significantly induced myeloid expansion in combination (Shi and Leung, unpublished), supporting the cooperativity of IDH and FLT3 mutations in AML leukemogenesis that has been reported in mouse models.44
Finally, the study has addressed the issues pertaining to the roles of zidh2 during embryonic development. zidh1 and zidh2 exhibited distinct spatial expression pattern at the embryonic stage and nonredundancy as shown by the incomplete rescue of pu.1 expansion in zidh1 embryos by zidh2 mRNA and the lack of effects of the latter on definitive hematopoiesis. However, knockdown of both zidh1 and zidh2 induced a similar increase in early myeloid progenitor and decrease in late myeloid/macrophage differentiation, suggesting the 2 genes might have a similar function in the regulation of myeloid differentiation, but only zidh1 had an effect on definitive hematopoiesis.
The observations in the study have provided important grounds for further investigations. For instance, IDH1 mutation and the resultant increase in 2-HG have been associated with suppression of TET2 that converts methylcytosine to hydroxymethylcytosine.45,46 Therefore, demonstration of a global increase in methylation in the developing embryos upon IDH1-R132H expression would provide a mechanistic link to the myeloid expansion and prove the principle that the pathogenetic pathways leading to leukemogenesis are conserved in the zebrafish embryos. Furthermore, cooperativity and mutual exclusivity between IDH1-R132H and other recurrent mutations in AML that have hitherto been technically challenging and cumbersome in mouse models could now be examined easily in the zebrafish model. These experiments will provide important insights to our understanding of the complex leukemogenesis in AML.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Professors Tak W. Mak, Steve C. Ekker, Racheal Wong, and Feng Liu for the helpful comments on the data.
This work was supported by the General Research Fund (HKU771110 and HKU729809M), the Collaborative Research Fund (HKUST5/CRF/12R), the National Natural Science Foundation of China (NSFC 81328004), the basic key research program from the Ministry of Science and Technology (2011CB964801) (T.C.), the Innovative Collaborative Research Programme and Zebrafish Core Facility at the Li Ka Shing Faculty of Medicine. A.Y.H.L. is the Li Shu Fan Medical Foundation Professor in Haematology and received funding from the endowment.
Authorship
Contribution: X.S. designed the study, performed some of the experiments, analyzed the data, and wrote the manuscripts; B-L.H., Y.G., Y.C., C.H.M., and A.C.H.M. performed some of the experiments; W.Z., Y.Z., Z.W., and T.C. designed the study and revised the manuscript; and A.Y.H.L. designed the study, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anskar Y. H. Leung, Room K418, K Block, Department of Medicine, Queen Mary Hospital, Pok Fu Lam Road, Hong Kong; e-mail: ayhleung@hku.hk.