Key Points
Increased quiescence of HSCs and HPCs in leukemogenesis, and reversible suppression of HSCs was observed in leukemic bone marrow.
A novel inhibitory role of Egr3 in HSC proliferation was revealed by leukemic infiltration in bone marrow.
Abstract
Cytopenias resulting from the impaired generation of normal blood cells from hematopoietic precursors are important contributors to morbidity and mortality in patients with leukemia. However, the process by which normal hematopoietic cells are overtaken by emerging leukemia cells and how different subsets of hematopoietic stem cells (HSCs) and hematopoietic progenitor cells (HPCs) are distinctly influenced during leukemic cell infiltration is poorly understood. To investigate these important questions, we used a robust nonirradiated mouse model of human MLL-AF9 leukemia to examine the suppression of HSCs and HPCs during leukemia cell expansion in vivo. Among all the hematopoietic subsets, long-term repopulating HSCs were the least reduced, whereas megakaryocytic-erythroid progenitors were the most significantly suppressed. Notably, nearly all of the HSCs were forced into a noncycling state in leukemic marrow at late stages, but their reconstitution potential appeared to be intact upon transplantation into nonleukemic hosts. Gene expression profiling and further functional validation revealed that Egr3 was a strong limiting factor for the proliferative potential of HSCs. Therefore, this study provides not only a molecular basis for the more tightened quiescence of HSCs in leukemia, but also a novel approach for defining functional regulators of HSCs in disease.
Introduction
The balance between primitive and mature blood cells is governed by both intrinsic1 and extrinsic factors.2,3 However, this balance can be severely disrupted in disease conditions, such as leukemia. Although normal hematopoietic cell proliferation, differentiation, and malignant transformation have been extensively investigated,4-6 the mechanisms by which normal hematopoietic cells are overcome by emerging leukemia cells in vivo and different subsets of hematopoietic stem cells (HSCs) and hematopoietic progenitor cells (HPCs) are distinctly influenced are poorly understood. Our previous work demonstrated that normal HSCs and HPCs were progressively suppressed during leukemia development but that they remained highly functional after being transplanted into nonleukemic recipients.7 This observation was consistent with a recent study showing the impact of human acute myeloid leukemia (AML) cells on HSCs in nonobese diabetic and severe combined immunodeficiency mice.8 There is also experimental evidence from other studies showing that leukemia cells can hijack the normal hematopoietic niche and create a leukemic microenvironment or directly change the bone marrow (BM) microenvironment to suppress the normal function of HSCs.9-11
The above studies are informative, as they have shown that normal HSCs and HPCs are suppressed in leukemia; however, unresolved issues preclude us from better understanding the response of normal hematopoietic cells to leukemia cell infiltration during disease development and more importantly, the mechanisms underlying the suppression of normal hematopoiesis. Thus, there is a need for an in vivo model that mimics the evolution of leukemia cells in patients and involves minimal experimental manipulations, such as immunosuppressive agents, total body irradiation (TBI), or xenotransplantation. TBI itself can destroy the immune system and normal HSC and HPC populations in recipients. Consequently, it exerts a significant bystander effect on transplanted cells in marrow.12 Thus, transplant protocols involving the use of TBI do not accurately reflect the conditions in leukemia patients. In addition, previous studies have focused on only one or a few HSC/HPC subsets, and they lacked data on the impact of leukemic hosts on the whole spectrum of different subsets of HSCs and HPCs in vivo. This issue is important because not all HSC and HPC subsets contribute equally to hematopoietic reconstitution after damage or transplantation. Moreover, an explanation of the molecular basis underlying the suppression of normal HSCs and HPCs is lacking. Therefore, an improved leukemia model may enable us to identify novel functional genes in HSCs under disease conditions, some of which have not been identified under normal homeostatic conditions.
This study used a robust nonirradiated acute leukemia mouse model, namely the MLL-AF9-induced AML model, to examine the kinetics of hematopoietic cell populations (including mature blood cell populations and different subsets of HSCs and HPCs) during leukemia cell infiltration in vivo. Distinct responses of different subsets of hematopoietic cells were observed. In particular, our results confirmed that HSCs were suppressed in leukemic BM and preserved in a noncycling state in the late stages of leukemia. Moreover, we identified a novel function of Egr3, a transcription factor, as a potent inhibitor of HSC proliferation due to leukemic cell infiltration in BM.
Methods
Mice
B6-Ly5.2 and B6-Ly5.1 mice were purchased from The Jackson Laboratory and maintained at the animal facility of the Institute of Hematology. The experimental protocol was approved by the Institutional Animal Care and Use Committee of the Institute of Hematology.
Flow cytometry
All the antibodies were obtained from BD Biosciences or e-Bioscience, unless otherwise specified. Staining was performed as previously described.13,14 The use of 4′,6 diamidino-2-phenylindole (DAPI; Sigma-Aldrich) excluded the dead cells. Detailed methodology can be found in the supplemental Methods on the Blood Web site.
Retroviral production
For retroviral production, 7 µg of the retroviral plasmid, 5 µg of pKat, and 3 µg of pVSVG were transfected into 293T cells using Lipofectamine 2000. After 48 and 72 hours of culture, the supernatant was harvested and concentrated using an Amicon filter.
Colony-forming assays
CD45.1+green fluorescent protein (GFP)− cells were isolated from the leukemia and control groups at different time points and then placed in MethoCult complete medium (M3434, StemCell Technologies). The cells were then plated in 24-well plates in 0.5 mL at 2 × 104 cells/mL. Colony-forming cells (CFCs) were scored after 10 days of incubation.
Leukemia mouse model
To establish a nonirradiated mouse model, 1 × 106 spleen cells (CD45.2+) harvested from primary leukemic mice or 1 × 106 normal BM cells (CD45.2+, control) were injected into nonirradiated recipients (CD45.1+).
Transplantation
Transplantations were performed as previously described.13 Detailed methodology can be found in the supplemental Methods.
Microarray analysis
CD45.1+Lin−c-Kit+ Sca-1+ (LKS+) cells were sorted from the leukemia and control groups at different time points. The microarray was performed at CapitalBio (Beijing, China). Gene set enrichment analysis was performed as previously described.15 HSC proliferative and quiescence gene sets were curated from a reference.16 Gene ontology (GO) and pathway analyses were performed at Genminix Informatics Ltd. (Shanghai, China). The microarray data have been deposited in NCBI’s Gene Expression Omnibus under accession number GSE52506.
Quantitative RT-PCR
Total RNA was isolated using a Qiagen RNeasy Mini Kit. cDNA was synthesized using Improm-II Reverse Transcriptase (Promega) or SuperScript III (Invitrogen). Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed with the primers or probes listed in supplemental Tables 2-3 using a 7500 or a StepOne Real-Time PCR System (Applied Biosystems).
Statistical analysis
GraphPad Prism 5.0 software was used for the statistical analysis. Unpaired Student t tests and analysis of variance were used to generate P values for most of the datasets.
Additional methods can be found in the supplemental Methods.
Results
Normal HSCs and HPCs progressively decreased in nonablated leukemic BM
To study the intact hematopoietic system, we established a nonirradiated AML model that is initiated by the human MLL-AF9 fusion protein17 (Figure 1A and supplemental Figure 1A). All the mice succumbed to leukemia with a median latency of 22 days (supplemental Figure 1B). The mice exhibited splenomegaly (supplemental Figure 1C), elevated white blood cell counts, mild anemia, and thrombocytopenia in the peripheral blood (PB) (supplemental Figure 1D). Leukemia progression was identified first in the BM and then in the spleen and PB; the percentage of CD45.1+ cells declined dramatically, whereas, the GFP+ leukemia cell population increased rapidly during leukemia development in all these tissues (supplemental Figure 1E). Thus, our results show that overall normal hematopoiesis is progressively suppressed in nonirradiated (nonmanipulated) mice during leukemia cell expansion.
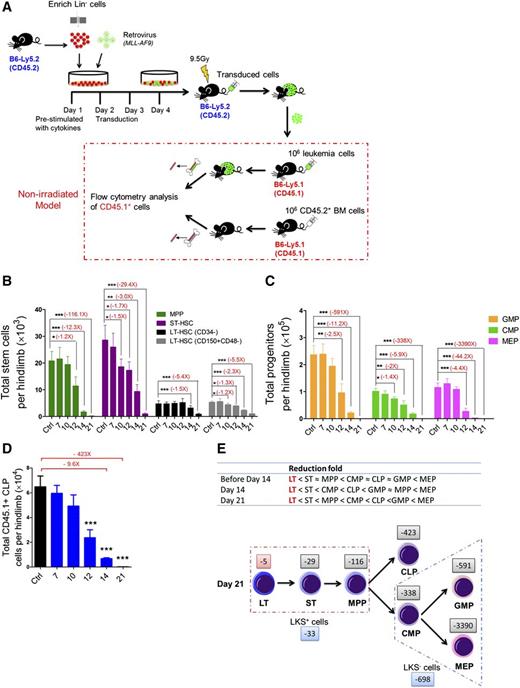
Kinetics of hematopoiesis in leukemic BM. (A) Retroviral transduction procedure and induction of leukemia in nonirradiated recipient mice. (B) Absolute numbers of LT-HSCs, ST-HSCs, and multipotent progenitors in leukemic BM. (C-D) Absolute numbers of GMPs, CMPs, MEPs (D), and CLPs (E) in leukemic BM. Data are represented as the mean ± standard error of the mean (SEM) (n = 12; 3 independent experiments). (E) Reduced speed (upper panel) and a model (lower panel) showing the differentiation block from HSCs to HPCs. The numbers (eg, −5 and −29) indicate the fold decrease. Plus (+) or minus (−), increase or decrease. *P < .05; **P < .01; ***P < .001.
Kinetics of hematopoiesis in leukemic BM. (A) Retroviral transduction procedure and induction of leukemia in nonirradiated recipient mice. (B) Absolute numbers of LT-HSCs, ST-HSCs, and multipotent progenitors in leukemic BM. (C-D) Absolute numbers of GMPs, CMPs, MEPs (D), and CLPs (E) in leukemic BM. Data are represented as the mean ± standard error of the mean (SEM) (n = 12; 3 independent experiments). (E) Reduced speed (upper panel) and a model (lower panel) showing the differentiation block from HSCs to HPCs. The numbers (eg, −5 and −29) indicate the fold decrease. Plus (+) or minus (−), increase or decrease. *P < .05; **P < .01; ***P < .001.
Next, we quantified the pool size of HSCs and HPCs at different time points in the BM after inoculating nonmanipulated recipients with leukemia cells. The frequencies of long-term HSCs (LT-HSCs) and short-term HSCs (ST-HSCs) in the CD45.1+ population gradually increased, whereas the frequency of multipotent progenitors (MPPs) was unchanged18-21 (supplemental Figure 2A-B). The absolute numbers of cells in all 3 subsets progressively decreased during leukemia development (Figure 1B). We also performed flow analysis of HPCs in leukemic BM: common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), megakaryocytic-erythroid progenitors (MEPs),22 and common lymphoid progenitors (CLPs).23 At day 21, the frequencies of all 4 subfractions were dramatically decreased (supplemental Figure 2A,C-D). The populations of all 4 progenitor subsets progressively decreased during leukemia development (Figure 1C-D). Notably, among all the subsets of primitive cells, LT-HSCs were the least reduced, whereas the MEPs were the most reduced at different stages of leukemia (Figure 1E). Thus, this result suggests that leukemic BM may affect normal hematopoietic differentiation kinetics, leading to a blockade of HSCs differentiation into HPCs, particularly MEPs in BM.
HSCs and HPCs in leukemic BM gradually accumulated in the G0 phase of the cell cycle
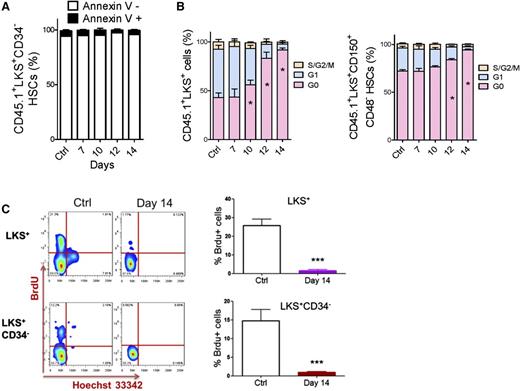
We performed apoptotic and cell cycle analyses to investigate how HSCs were suppressed. The ratio of Annexin V+ cells in HSC populations was nearly unchanged during leukemia development (Figure 2A and supplemental Figure 3A). However, the proportion of HSCs in G0 significantly increased during leukemia development (Figure 2B and supplemental Figure 3B). Moreover, HSCs in the late stages (after day 14) of leukemia remained in a nearly nonproliferative state (95% vs 70% of HSCs were in G0). This result was confirmed by a 5-bromo-2′-deoxyuridine (BrdU) incorporation assay (Figure 2C). Therefore, the suppression of HSCs in leukemic BM was mainly due to a functional alteration in the cell cycle, not apoptosis.
Apoptosis and cell cycle status of HSCs in leukemic BM. (A) Apoptosis rate of HSCs in leukemic BM. (B) Cell cycle status of HSCs in leukemic BM. (C) Flow plots (left panel) and histogram (right panel) show the BrdU incorporation of HSCs (LKS+ and LKS+CD34−) in leukemic BM. Data are represented as the mean ± SEM (n = 12; 3 independent experiments). Ctrl, control. *P < .05; *** P < .001.
Apoptosis and cell cycle status of HSCs in leukemic BM. (A) Apoptosis rate of HSCs in leukemic BM. (B) Cell cycle status of HSCs in leukemic BM. (C) Flow plots (left panel) and histogram (right panel) show the BrdU incorporation of HSCs (LKS+ and LKS+CD34−) in leukemic BM. Data are represented as the mean ± SEM (n = 12; 3 independent experiments). Ctrl, control. *P < .05; *** P < .001.
Similar to the HSC findings, there was no significant difference in the apoptotic fraction in HPCs (LKS−) in the leukemic BM and cells from the control BM (supplemental Figure 4A). This result also indicates that apoptosis is not responsible for the decreased number of HPCs. However, the proportion of HPCs in G0 increased significantly after day 10 (supplemental Figure 4B), and more HPCs remained in a nonproliferative state at day 14 (supplemental Figure 4C). After HSC cycling ceases, HPCs cannot be replenished in leukemic BM. However, HPCs need to produce mature blood cells. Therefore, unlike the behavior of HSCs, the number of HPCs sharply declined, and this population was ultimately exhausted. Only a small fraction of HPCs was observed to be in a quiescent state.
Normal HSCs and HPCs in leukemic spleen were relatively active
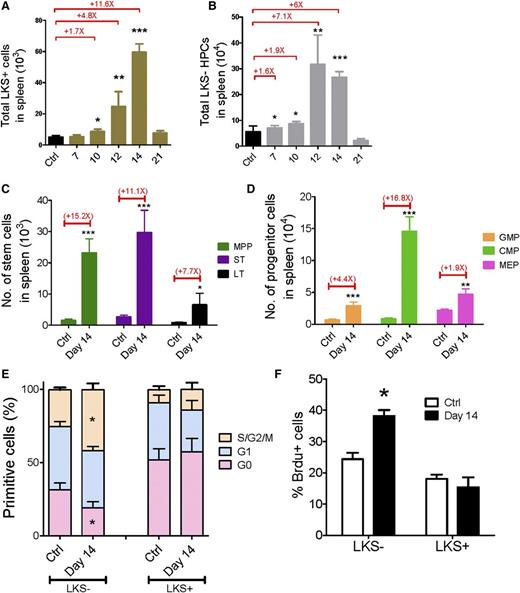
To understand whether extramedullary hematopoiesis occurs in the spleen, we analyzed the changes in HSC and HPC populations in the leukemic spleen. Both the frequencies and absolute numbers of LKS+ and LKS− cells in the leukemic spleen increased from day 7 to day 14 and then dropped to normal levels at day 21 (Figure 3A-B and supplemental Figure 5A-B). Moreover, we quantified the changes in the different subsets of primitive cells at leukemia day 14: both the frequencies and absolute numbers of all subsets in the spleen were increased (Figure 3C-D and supplemental Figure 5C-D). Thus, these data suggest that unlike leukemic BM, the leukemic spleen does not suppress hematopoietic cells.
Kinetics of hematopoiesis in the leukemic spleen. (A-B) Absolute numbers of LKS+ (A) and LKS− (B) cells in the leukemic spleen. (C-D) Absolute numbers of LT-HSCs, ST-HSCs, MPPs (C), GMPs, CMPs, and MEPs (D) in the leukemic spleen. (E) Ki67 staining of LKS+ and LKS− cells in the leukemic spleen. (F) BrdU staining of LKS+ and LKS− cells in the leukemic spleen. Data are represented as the mean ± standard error of the mean (n = 12; 3 independent experiments). Ctrl, control. *P < .05; **P < .01; ***P < .001.
Kinetics of hematopoiesis in the leukemic spleen. (A-B) Absolute numbers of LKS+ (A) and LKS− (B) cells in the leukemic spleen. (C-D) Absolute numbers of LT-HSCs, ST-HSCs, MPPs (C), GMPs, CMPs, and MEPs (D) in the leukemic spleen. (E) Ki67 staining of LKS+ and LKS− cells in the leukemic spleen. (F) BrdU staining of LKS+ and LKS− cells in the leukemic spleen. Data are represented as the mean ± standard error of the mean (n = 12; 3 independent experiments). Ctrl, control. *P < .05; **P < .01; ***P < .001.
Then we extended the cell cycle analysis to the spleen LKS+ and LKS− cells at day 14 to determine the proliferative rate of normal hematopoietic cells. Interestingly, Ki67 staining revealed that only 19% of LKS− cells from leukemic spleen remained in G0 compared with 31% of LKS− cells from the control spleen, whereas the cell cycle status of the LKS+ cells from the leukemia and control spleens was comparable (Figure 3E). A similar result was observed in the BrdU incorporation assay (Figure 3F). LKS− HPCs from the leukemic spleen exhibited greater BrdU labeling than those from the control spleen (38% vs 24%, respectively), whereas no significant difference in BrdU incorporation was observed in the LKS+ cells (Figure 3F). These data indicate that the HPCs demonstrate an increased proliferative rate and the LKS+ cells show a normal cell cycle profile in the leukemic spleen, which suggests that there is increased hematopoiesis activity in the leukemic spleen, and the spleen may compensate for hematopoietic suppression in leukemic BM, at least in certain types of leukemia (eg, MLL-AF9 AML).
HSC function was preserved in leukemic BM
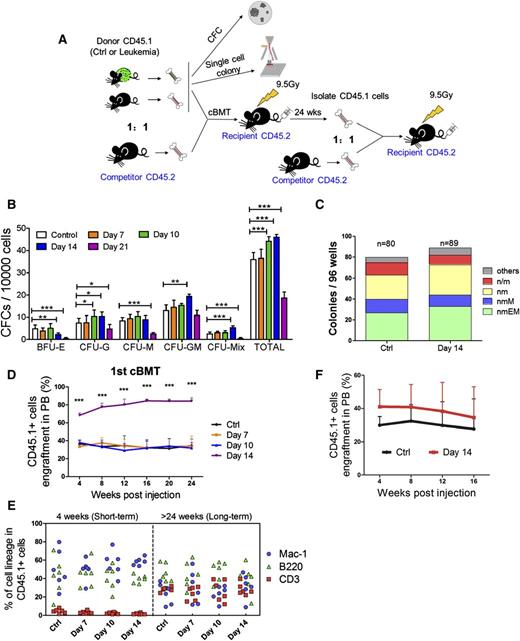
To gauge the functional alteration of HSCs and HPCs in the leukemic environment, we performed an in vitro CFC assay (Figure 4A) first. CD45.1+ cells from leukemia day 14 triggered significantly more colony-forming unit (CFU)-G, CFU-GM, and CFU-mix colonies, but much fewer burst-forming unit erythroid colonies. Strikingly, the frequencies of all colony types were significantly lower at leukemia day 21 compared with the control (Figure 4B). These changes are partially consistent with our phenotypic analysis, which showed that the frequency of HPCs transiently increased from day 10 to day 14, and it showed that the HPC population was exhausted by day 21 (supplemental Figures 2A and 6A). A single-cell liquid culture assay was also performed to compare the in vitro multilineage proliferation of LKS+CD34− LT-HSCs from the leukemic (day 14) and control mice (Figure 4A).14 Overall, 83% to 92% of the cells formed colonies, and the frequency of each colony type was similar between the 2 groups (Figure 4C), suggesting that the differentiation potential was not significantly altered in HSCs isolated from leukemic marrow.
Functional assessments of hematopoietic cells isolated from leukemic mice. (A) Scheme of the colony assays and cBMTs. (B) The histogram shows the colony-forming ability of normal hematopoietic cells from leukemic and control BM. Data are represented as the mean ± SEM (n = 12; 2 independent experiments). (C) The histogram shows the single cell colony-forming ability of normal LT-HSCs from the leukemic and control BM. (D) The percentage of CD45.1+ donor cells in the PB of recipient mice at the indicated time points after primary cBMT. The donor cells were CD45.1+ BM cells. Data are represented as the mean ± SEM (n = 10-12; 2 independent experiments). (E) Short-term and long-term multilineage reconstitution capacities of donor-derived (CD45.1+) PB cells in primary recipients. (F) The percentage of CD45.1+ donor cells in the PB of recipient mice at the indicated time points after primary cBMT. Donor cells were CD45.1+LKS+CD34− LT-HSCs. Data are represented as the mean ± standard deviation (SD) (n = 7). Ctrl, control. *P < .05; **P < .01; ***P < .001.
Functional assessments of hematopoietic cells isolated from leukemic mice. (A) Scheme of the colony assays and cBMTs. (B) The histogram shows the colony-forming ability of normal hematopoietic cells from leukemic and control BM. Data are represented as the mean ± SEM (n = 12; 2 independent experiments). (C) The histogram shows the single cell colony-forming ability of normal LT-HSCs from the leukemic and control BM. (D) The percentage of CD45.1+ donor cells in the PB of recipient mice at the indicated time points after primary cBMT. The donor cells were CD45.1+ BM cells. Data are represented as the mean ± SEM (n = 10-12; 2 independent experiments). (E) Short-term and long-term multilineage reconstitution capacities of donor-derived (CD45.1+) PB cells in primary recipients. (F) The percentage of CD45.1+ donor cells in the PB of recipient mice at the indicated time points after primary cBMT. Donor cells were CD45.1+LKS+CD34− LT-HSCs. Data are represented as the mean ± standard deviation (SD) (n = 7). Ctrl, control. *P < .05; **P < .01; ***P < .001.
To stringently test whether leukemia affects HSC function in vivo, we performed competitive BM transplantation (cBMT) (Figure 4A). The repopulation ability of BM cells from days 7 and 10 leukemic BM was similar to that of the cells from the control BM (Figure 4D). Surprisingly, the BM cells from day 14 mice exhibited significantly higher reconstitution than the control cells (82% vs 35%). Donors (CD45.1+) from the 4 groups had the same capacity to generate myeloid (Mac-1+) and lymphoid (CD3+ and B200+) lineages (Figure 4E and supplemental Figure 6B). Similar results were obtained by analyzing the BM (supplemental Figure 6C-E). Together, these results indicate that there are no defects in the reconstitution potential of the residual normal HSCs from the leukemic environment.
The frequency of LT-HSCs in day 14 leukemic BM was 7.1-fold higher than that in the control BM, and the CD45.1+ cells from day 14 leukemic BM exhibited 7.4-fold higher donor cell engraftment at 24 weeks posttransplantation (supplemental Figure 2B and 6C). These data may explain the increased engraftment in the recipients when unseparated BM cells were transplanted. To confirm this hypothesis, LT-HSCs were isolated from leukemia and control BM at day 14 and then used as donors in a cBMT assay. As expected, LT-HSCs from the leukemic BM showed a small, but not significant increase in long-term repopulating activity (Figure 4F). Together, these data indicate that the reconstitution potential of HSCs in the leukemic mice and the control mice is equal (average function per HSC), which was consistent with the results from the single-cell liquid culture assay (Figure 4C). Secondary competitive transplantation showed no further significant difference in repopulating activity (supplemental Figure 6F-G), suggesting that the self-renewal capacity of HSCs was restored. Thus, our data indicate that although leukemic stress suppresses normal HSCs by forcing them into a noncycling state, HSC function is well preserved.
HSCs from leukemic BM had a distinct gene expression signature
To investigate the molecular basis for the reversible suppression of HSCs by leukemia, we sorted the CD45.1+LKS+ hematopoietic stem and progenitor cells (HSPCs) from control or leukemic mice at days 7 and 14 for gene expression profiling analysis (Figure 5A). In agreement with the cell cycle analysis, gene set enrichment analysis revealed that the expression of quiescence-associated genes was significantly enriched in the LKS+ cells from the day 14 leukemic mice, whereas the expression of proliferation-associated genes was markedly downregulated (Figure 5B). GO and pathway analyses both indicated that the downregulated genes were highly enriched for “cell cycle” catalogs, and the cell cycle pathway was suppressed (supplemental Figure 7A-C). Then we confirmed the expression of the cell cycle-related genes (CKIs, CDKs, and cyclins) that were identified in the microarray data through qRT-PCR. p19INK4d and p21Cip1 were significantly upregulated in the LKS+ cells from the leukemic BM (Figure 5C and supplemental Figure 7D). Most of the CDKs and cyclins were significantly downregulated in the LKS+ cells from the leukemic mice (Figure 5D-E and supplemental Figure 7D). These data reveal that leukemic stress forced the HSCs to remain quiescent by suppressing cell cycle-promoting gene expression and enhancing cell cycle-inhibitory gene expression.
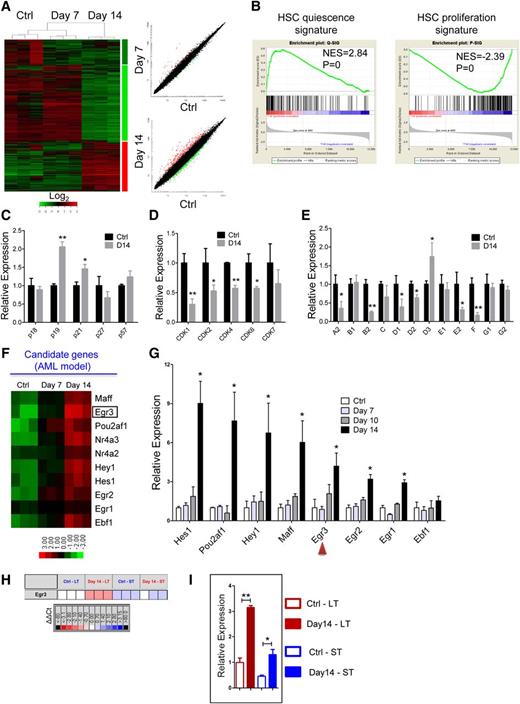
Differential gene expression of HSCs from the leukemic and control mice. (A) Heatmap (left panel) and scatter-plot representation (right panel) of differential gene expression in normal LKS+ cells at different stages of leukemia. The color scale indicates normalized expression values. (B) Gene set enrichment analysis comparison of LKS+ cells from day 14 leukemia and control mice: the upregulation or downregulation of HSC quiescence-associated gene expression (left panel) and proliferation-associated gene expression (right panel). The normalized enrichment scores (NES) and P values are indicated in each plot. (C-E) The histograms show cell cycle-related gene expression levels in BM LKS+ cells at leukemia day 14 compared with the control. CKIs (C), CDKs (D), and cyclins (E). Data are represented as the mean ± SEM (n = 9; 3 independent experiments). (F) Heatmap representation of candidate gene expression in normal LKS+ cells at different stages of AML. The color scale indicates normalized expression values. (G) The qRT-PCR analysis shows the expression levels of candidate genes in normal LKS+ cells at different stages of AML. Data are represented as the mean ± SEM (n = 9; 3 independent experiments). The red arrowheads indicate the 2 genes that we studied further. (H) The quantitative polymerase chain reaction array analysis of Egr3 expression in LT-HSCs (LKS+CD34−) and ST-HSCs (LKS+CD34+) from day 14 leukemia and control mice. Representative heat map of δ-δ Ct (ΔΔCt) values of positive signals. The red and blue colors indicate high and low gene expression, respectively, relative to the reference. Black indicates no detectable signal. (I) Egr3 expression in LT-HSCs and ST-HSCs from day 14 leukemia and control mice (refer to Figure 5H). Data are represented as the mean ± SEM. Ctrl, control. *P < .05; **P < .01.
Differential gene expression of HSCs from the leukemic and control mice. (A) Heatmap (left panel) and scatter-plot representation (right panel) of differential gene expression in normal LKS+ cells at different stages of leukemia. The color scale indicates normalized expression values. (B) Gene set enrichment analysis comparison of LKS+ cells from day 14 leukemia and control mice: the upregulation or downregulation of HSC quiescence-associated gene expression (left panel) and proliferation-associated gene expression (right panel). The normalized enrichment scores (NES) and P values are indicated in each plot. (C-E) The histograms show cell cycle-related gene expression levels in BM LKS+ cells at leukemia day 14 compared with the control. CKIs (C), CDKs (D), and cyclins (E). Data are represented as the mean ± SEM (n = 9; 3 independent experiments). (F) Heatmap representation of candidate gene expression in normal LKS+ cells at different stages of AML. The color scale indicates normalized expression values. (G) The qRT-PCR analysis shows the expression levels of candidate genes in normal LKS+ cells at different stages of AML. Data are represented as the mean ± SEM (n = 9; 3 independent experiments). The red arrowheads indicate the 2 genes that we studied further. (H) The quantitative polymerase chain reaction array analysis of Egr3 expression in LT-HSCs (LKS+CD34−) and ST-HSCs (LKS+CD34+) from day 14 leukemia and control mice. Representative heat map of δ-δ Ct (ΔΔCt) values of positive signals. The red and blue colors indicate high and low gene expression, respectively, relative to the reference. Black indicates no detectable signal. (I) Egr3 expression in LT-HSCs and ST-HSCs from day 14 leukemia and control mice (refer to Figure 5H). Data are represented as the mean ± SEM. Ctrl, control. *P < .05; **P < .01.
GO analysis indicated that the upregulated genes were highly enriched for “transcription” catalogs (supplemental Figure 7E). Ten transcription-related genes, which were highly expressed in the LKS+ cells in leukemic BM, were selected as candidates (Figure 5F, supplemental Table 1). Certain selected genes, such as Egr1,24,25 Hes1,26,27 and Nr4a228 were previously implicated in the regulation of HSCs, and some genes were not known to be associated with HSC function. Eight genes were validated by qRT-PCR in LKS+ cells (Figure 5G). Egr3 is an early response gene belonging to a family of zinc-finger transcription factors that also includes Egr1, Egr2, and Egr4. Egr1 controls HSC proliferation,24 whereas Egr2 and Egr3 appear to be important for lymphocyte proliferation or activation29,30 ; Egr1/2/3 were all highly expressed in the LKS+ cells in the leukemic BM. Collectively, these results suggest a potential role for Egr3 in HSCs, which prompted us to further investigate its function in HSCs.
Next, we used Fluidigm quantitative polymerase chain reaction analysis to determine whether Egr3 was also highly expressed in LKS+CD34− LT-HSCs from leukemic BM. Egr3 expression was more than 3-fold (or 2-fold) higher in the LT-HSCs (or ST-HSCs) from leukemic BM than in the LT-HSCs (or ST-HSCs) from control BM (Figure 5H-I). We further determined the expression abundance of Egr3 at the single cell level: Egr3 was highly expressed in the LT-HSCs and ST-HSCs from leukemic BM compared with control BM (supplemental Figure 8).
Egr3 expression correlated with the cell proliferative state
In performing a cBMT assay, we found that the expression level of Egr3 decreased in LKS+ cells when they were transplanted into nonleukemic hosts (Figure 6A), and that the cell cycle status of LKS+ cells normalized when they were released from leukemic stress (Figure 6B). Furthermore, the HSPCs from the leukemic spleen showed normal cell cycle distribution (Figure 3E-F) and normal Egr3 expression levels (Figure 6C). These data demonstrated that leukemic BM stress increased Egr3 expression in normal HSPCs and that Egr3 expression was related to cell cycle status.
Correlation of Egr3 expression with HSPC proliferation. (A) Egr3 expression in normal LKS+ cells in leukemic and nonleukemic microenvironments. Normal cells (CD45.1) were isolated from leukemia day 14 and transplanted into nonleukemic recipients. LKS+ cells were sorted 1 to 4 weeks after reconstitution. Data are represented as the mean ± SD (n = 3). (B) Flow plots (left panel) and histogram (right panel) of the cell cycle status of donor LKS+ cells from primary recipients (control [Cntrl] vs day 14). Data are represented as the mean ± SD (n = 6). (C) Egr3 expression in LKS+ cells from leukemic and control spleen. Data are represented as the mean ± SD (n = 6). (D) Coculture of LKS+ cells with BM plasma. Normal LKS+ cells were cultured with BM plasma obtained from control and day 14 leukemic mice. The proliferation of LKS+ cells was markedly suppressed by the leukemic BM plasma. Data are represented as the mean ± SEM (n = 6; 2 independent experiments). (E) LKS+ cells were treated as indicated in Figure 6D. Egr3 expression is shown. Data are represented as the mean ± SEM (n = 6; 2 independent experiments). **P < .01; ***P < .001.
Correlation of Egr3 expression with HSPC proliferation. (A) Egr3 expression in normal LKS+ cells in leukemic and nonleukemic microenvironments. Normal cells (CD45.1) were isolated from leukemia day 14 and transplanted into nonleukemic recipients. LKS+ cells were sorted 1 to 4 weeks after reconstitution. Data are represented as the mean ± SD (n = 3). (B) Flow plots (left panel) and histogram (right panel) of the cell cycle status of donor LKS+ cells from primary recipients (control [Cntrl] vs day 14). Data are represented as the mean ± SD (n = 6). (C) Egr3 expression in LKS+ cells from leukemic and control spleen. Data are represented as the mean ± SD (n = 6). (D) Coculture of LKS+ cells with BM plasma. Normal LKS+ cells were cultured with BM plasma obtained from control and day 14 leukemic mice. The proliferation of LKS+ cells was markedly suppressed by the leukemic BM plasma. Data are represented as the mean ± SEM (n = 6; 2 independent experiments). (E) LKS+ cells were treated as indicated in Figure 6D. Egr3 expression is shown. Data are represented as the mean ± SEM (n = 6; 2 independent experiments). **P < .01; ***P < .001.
Thus, we isolated LKS+ cells from normal B6-Ly5.1 mice and treated these cells with BM plasma from the leukemic and control mice. Compared with control BM plasma, the leukemic BM plasma suppressed the proliferation of HSPCs (Figure 6D) and unregulated Egr3 expression in the HSPCs (Figure 6E), demonstrating that Egr3 can be induced under leukemic conditions and the upregulation of Egr3 suppressed HSPC proliferation.
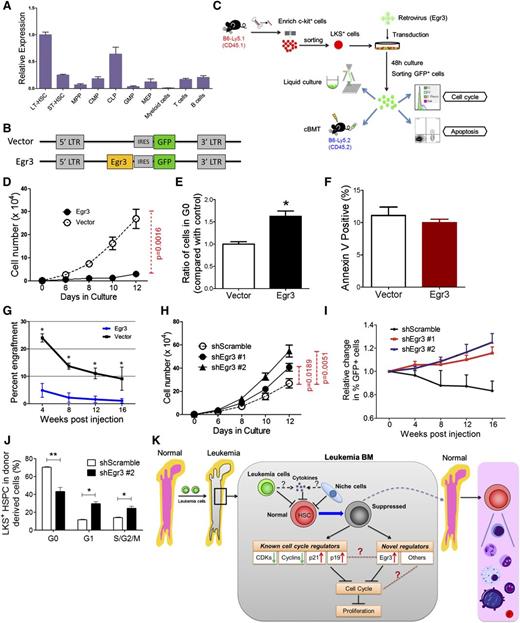
Egr3 was a strong limiting factor for the proliferative potential of mouse HSCs
As evidenced by qRT-PCR, Egr3 was specifically and highly expressed in mouse LT-HSCs (Figure 7A). Because Egr3 was highly expressed in the HSCs from the leukemic BM (Figure 5G), we forced its expression in LKS+ cells to determine whether its overexpression in the HSPCs was functionally associated with the suppression of proliferation. The cells were transduced with a retrovirus carrying a vector containing Egr3 upstream of an internal ribosome entry site (IRES)-GFP (Figure 7A-C). Meanwhile, an identical vector carrying only GFP was used as a control.
Functional impact of Egr3 expression in HSPCs. (A) The gene expression pattern of Egr3 in hematopoietic cells at different stages. Data are represented as the mean ± SD (n = 3). (B) Diagram of the vectors used to express Egr3 in LKS+ cells. Egr3 cDNA was cloned into murine stem cell virus for subsequent transduction into cells. (C) Schematic of the overexpression experiments in LKS+ cells using the indicated retroviruses shown in Figure 7B. (D) LKS+ cells were treated as indicated in Figure 7C. GFP+ cell proliferation is shown. (E) Quantification of cells remaining in G0, as indicated in Figure 7C. Data are represented as the mean ± SEM (n = 6-8; 2 independent experiments). (F) Apoptotic analysis of LKS+ cells 48 hours after transduction with control or Egr3 retrovirus. Data are represented as the mean ± SEM (n = 6-8; 2 independent experiments). (G) The percentage of GFP+ donor cells in the PB of recipient mice at the indicated time points. Data are represented as the mean ± SD (n = 7-9). (H) In vitro liquid culture of Egr3-knockdown cells. Data are represented as the mean ± SEM (n = 8; 2 independent experiments). (I) The histogram shows changes in PB chimerism of GFP+ cells in recipients at the indicated time points after transplantation. In this assay, we did not sort the GFP+ cells after 48 hours of transduction. We directly injected the GFP+ and GFP− cells into the recipients. The percentage of cells expressing GFP on the day of injection was normalized to 1. Data are represented as the mean ± SD (n = 6-8). (J) The cell cycle status of LKS+ cells in leukemic BM after Egr3 knockdown. Data are represented as the mean ± SEM (n = 8; 2 independent experiments). (K) Schematic of the response of HSCs to leukemia stress. The molecular mechanisms by which leukemia affects normal HSCs, as suggested by our studies, are indicated. IRES, internal ribosome entry site; LTR, long terminal repeats. *P < .05; **P < .01.
Functional impact of Egr3 expression in HSPCs. (A) The gene expression pattern of Egr3 in hematopoietic cells at different stages. Data are represented as the mean ± SD (n = 3). (B) Diagram of the vectors used to express Egr3 in LKS+ cells. Egr3 cDNA was cloned into murine stem cell virus for subsequent transduction into cells. (C) Schematic of the overexpression experiments in LKS+ cells using the indicated retroviruses shown in Figure 7B. (D) LKS+ cells were treated as indicated in Figure 7C. GFP+ cell proliferation is shown. (E) Quantification of cells remaining in G0, as indicated in Figure 7C. Data are represented as the mean ± SEM (n = 6-8; 2 independent experiments). (F) Apoptotic analysis of LKS+ cells 48 hours after transduction with control or Egr3 retrovirus. Data are represented as the mean ± SEM (n = 6-8; 2 independent experiments). (G) The percentage of GFP+ donor cells in the PB of recipient mice at the indicated time points. Data are represented as the mean ± SD (n = 7-9). (H) In vitro liquid culture of Egr3-knockdown cells. Data are represented as the mean ± SEM (n = 8; 2 independent experiments). (I) The histogram shows changes in PB chimerism of GFP+ cells in recipients at the indicated time points after transplantation. In this assay, we did not sort the GFP+ cells after 48 hours of transduction. We directly injected the GFP+ and GFP− cells into the recipients. The percentage of cells expressing GFP on the day of injection was normalized to 1. Data are represented as the mean ± SD (n = 6-8). (J) The cell cycle status of LKS+ cells in leukemic BM after Egr3 knockdown. Data are represented as the mean ± SEM (n = 8; 2 independent experiments). (K) Schematic of the response of HSCs to leukemia stress. The molecular mechanisms by which leukemia affects normal HSCs, as suggested by our studies, are indicated. IRES, internal ribosome entry site; LTR, long terminal repeats. *P < .05; **P < .01.
Flow cytometry analysis revealed that transduction efficiency after 48 hours in culture was 59% to 87% (supplemental Figure 9A). GFP+ cells were sorted and used for assays including liquid culture, CFC assays, and transplantation (Figure 7C). The qRT-PCR confirmed the Egr3 expression level in the GFP+ cells (supplemental Figure 9B). In vitro liquid culture revealed that Egr3 overexpression restricted the proliferation of GFP+ cells (P = .0016; Figure 7D) and led to an approximately 60% increase in the number of cells in G0 compared with the control (Figure 7E). Moreover, an Annexin V staining assay showed that there was no increase in cell death associated with Egr3 overexpression (Figure 7F). Thus, our data indicate that Egr3 inhibits the proliferation of HSPCs.
To determine the impact of Egr3 on HSC function, we transplanted transduced cells (GFP+) and wild-type CD45.2+ BM cells into lethally irradiated mice (B6-Ly5.2) (Figure 7C). Egr3-transduced cells contributed an extremely low number of cells to the reconstituted PB (Figure 7G). Noncompetitive transplantation was performed to confirm the negligible contribution of Egr3-overexpressing donor cells to PB reconstitution (supplemental Figure 9C-E). Using a complementary approach, we transduced LKS+ cells with lentiviruses carrying Egr3 short hairpin RNA (shRNA) to knockdown Egr3 expression. Flow cytometry analysis revealed that after 48 hours in culture, the transduction efficiency was approximately 90% (supplemental Figure 9F). The 2 shRNAs exhibited good knockdown efficiency at the mRNA level (supplemental Figure 9G). In vitro liquid culture showed that Egr3 knockdown promoted the proliferation of transduced cells (Figure 7H). Non-competitive and competitive transplantation demonstrated that Egr3 depletion improved the repopulation ability of the LKS+ cells (Figure 7I and supplemental Figure 9H). To determine whether Egr3 depletion could counteract the HSC quiescence in leukemic marrow, we transplanted Egr3 knockdown LKS+ cells into lethally irradiated recipients. After 17 days of reconstitution, we injected the AML cells to establish the non-irradiated leukemia model. At leukemia day 12, we analyzed the cell cycle of the normal LKS+ cells. Compared with the control, there were fewer quiescent cells among the Egr3-knockdown LKS+ HSPC population (Figure 7J). Together, these results indicate that Egr3 suppresses the function of HSCs and plays a significant role in silencing HSCs in leukemic marrow. Moreover, targeting Egr3 in HSCs in leukemia can rescue this suppression.
Discussion
In this study, we used an unmanipulated AML mouse model to measure the suppression of different HSC and HPC subsets in leukemic marrow, in which the HSCs were nearly synchronized to a quiescent state with full repopulating potential. Furthermore, through microarray analysis and functional validation, we demonstrated that Egr3 strongly restricted the proliferation of HSCs (Figure 7K), indicating its key role in silencing HSCs in BM under leukemic conditions.
Normal HSC suppression under leukemic stress is determined by both intrinsic and extrinsic regulators. Our current study mainly focused on changes in intrinsic regulators. Of the candidate genes identified by our microarray analysis (Figure 5F), the upregulated genes Egr1, Nr4a2, or Egr3 were suggested to strongly suppress HSC function, and Hes1 overexpression may potentiate HSC function. The distinct functions of Egr1, Nr4a2, and Hes1 in HSCs have been reported in previous studies.25,26,28 Although HSC function was suppressed and HSC proliferation was completely inhibited in the leukemic BM, the reconstitution and self-renewal capacities of HSCs were fully preserved and recovered under nonleukemic conditions, as previously documented in the irradiated leukemia model.7 Our current study further suggests a paradigm in which suppressor genes (such as Egr3) lead to the suppression of HSCs and potentiator genes (such as Hes1) contribute to the functional preservation of HSCs during leukemia development. Thus, the balance between suppressors and potentiators is critical for HSC function in leukemia. Previously, Egr3 has been shown to be a positive regulator of lymphocyte proliferation.30-32 Here, we defined Egr3 as a negative regulator of HSC proliferation and suggested that the specific effect of Egr3 on cell cycle appear to depend on the lineage/differentiation stage of affected cells. Moreover, its inhibitory effect was likely due to specific microenvironmental cues in the leukemic marrow as opposed to the spleen.
As to the extrinsic regulators, we found that compared with control plasma, the BM plasma from the leukemic hosts suppressed the proliferation of HSPCs (Figure 6D), suggesting that certain cytokines secreted by leukemia cells or microenvironmental cells in BM suppress HSC function (Figure 7K). Interestingly, our study also shows that the leukemic spleen does not exert a specific inhibitory effect on normal hematopoietic cells. In fact, the leukemic spleen was also able to promote leukemic cell homing and proliferation in comparison with leukemic BM.33 In addition, the potentially increased inflammatory cytokines cannot explain the increased number of quiescent HSCs during leukemia development, despite the fact that previous reports have demonstrated an increased cycling fraction of HSCs under infection stress.34,35 Our future studies will focus on (1) the extrinsic effects of leukemia on normal HSCs, including the alterations in cytokines and niche cells in leukemic BM, (2) how cytokine or niche cells regulate Egr3 expression in HSCs, and (3) the relationship between Egr3 and cell cycle regulatory genes.
Anemia and thrombopenia contribute to morbidity and mortality in patients with leukemia. This result can be partially explained by our finding of a dramatic reduction in MEPs, the upstream precursors of platelets and red cells (Figure 1E). Among the evaluated HSC and HPC subsets, MEPs were the most impacted, whereas LT-HSCs were the least impacted by the leukemic environment. Interestingly, in our parallel study, we found that normal MEPs were diminished in an acute graft-versus-host disease model.36 Because MEPs alone provide radioprotection,37 and the phenomenon has occurred in various disease and injury models, MEPs appear to play vital and unique roles in defective hematopoiesis under certain pathological conditions; thus, a special consideration may be given to the protection or augmentation of MEPs in the development of more effective regimens for at least some types of leukemia. As an extension, in light of the recent study showing the limited contribution of LT-HSCs to native hematopoiesis and the prolonged presence of some classical myeloid progenitor cells under homeostatic conditions,38 it will be important and exciting to evaluate the actual contributions of MEPs, as well as other different HSC and HPC subsets to hematopoiesis in different types of leukemia and in other diseases in the future.
In summary, this study not only documents the process of and mechanistic insights into the reversible suppression of HSCs in leukemic hosts, but it also provides new approaches and valuable resources for the future study of adult HSC biology in the context of disease, such as leukemia, thereby prompting the study of normal hematopoiesis in response to leukemia cell outgrowth.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr K. Lenhard Rudolph for providing the spleen-focus forming promoter-shRNA-GFP lentiviral vector, and Dr Qianfei Wang’s laboratory for analyzing the microarray data. The authors are grateful to their laboratory members and collaborators for assisting with the experiments and with the manuscript preparation. This work was supported by grants from the Ministry of Science and Technology of China (2011CB964801, 2012CB966600, 2010CB945204) and from the National Nature Science Foundation of China (81090411, 81421002, 81430004, 81130074, 81400077, 81300375, and 81300374).
Authorship
Contribution: H.C. and T.C. designed experiments; H.C. performed all experiments; H.C., G.Z., S.L., and W.Y. analyzed data; H.C. and T.C. wrote the manuscript; S.H., Y.L., and J.X. contributed to mouse experiments; S.H. contributed to data analysis; Y.P. helped with flow cytometry; S.M. and F.D. participated in single-cell quantitative polymerase chain reaction; S.L. contributed to microarray data analyses; G.Z., S.L., and W.Y. conducted research and assisted with manuscript preparation; and T.C. conceived the study, interpreted the results, and oversaw the research project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tao Cheng, 288 Nanjing Rd, Tianjin 300020, China; e-mail: chengtao@ihcams.ac.cn.
References
Author notes
H.C. and S.H. contributed equally to this work.

![Figure 6. Correlation of Egr3 expression with HSPC proliferation. (A) Egr3 expression in normal LKS+ cells in leukemic and nonleukemic microenvironments. Normal cells (CD45.1) were isolated from leukemia day 14 and transplanted into nonleukemic recipients. LKS+ cells were sorted 1 to 4 weeks after reconstitution. Data are represented as the mean ± SD (n = 3). (B) Flow plots (left panel) and histogram (right panel) of the cell cycle status of donor LKS+ cells from primary recipients (control [Cntrl] vs day 14). Data are represented as the mean ± SD (n = 6). (C) Egr3 expression in LKS+ cells from leukemic and control spleen. Data are represented as the mean ± SD (n = 6). (D) Coculture of LKS+ cells with BM plasma. Normal LKS+ cells were cultured with BM plasma obtained from control and day 14 leukemic mice. The proliferation of LKS+ cells was markedly suppressed by the leukemic BM plasma. Data are represented as the mean ± SEM (n = 6; 2 independent experiments). (E) LKS+ cells were treated as indicated in Figure 6D. Egr3 expression is shown. Data are represented as the mean ± SEM (n = 6; 2 independent experiments). **P < .01; ***P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/11/10.1182_blood-2015-01-623645/4/m_1302f6.jpeg?Expires=1763465261&Signature=qHXX8DZNpP6OMXKR48NnVOaeNogN79fQZpbgm4sjfEK2jbyNxVu7UHpwDYYE~sT4FRb0JpOU8Kx-N6n2V0btm5UwcbDx8GZhy9zZLknysoMHjB5qDsdRsVXnwRPyxCHVJTaAsYf82bpZMBy-Pwqxm-mLkIgg2BDXQEdJqHmsj~7Had1sEsYrka-Twxw5Rkui3NNzowA5vy9rUHtB7sNuZnnhUK9VsXqSNbgWeG2pPfeLr2hrbq0odDTzXWvHti-aVsjqX~TvIag~GisYNkF8CsEo6maVI3WznPpYsWunpVeNbLOCfbmnryoS-MlL92wIfmDqDTpNQty9RUERX5~wDQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal