Key Points
TP53 and RAS-pathway mutations predict very poor survival, when seen with CK and MDS/MPNs, respectively.
For patients with mutated TP53 or CK alone, long-term survival could be obtained with stem cell transplantation.
Abstract
Genetic alterations, including mutations and copy-number alterations, are central to the pathogenesis of myelodysplastic syndromes and related diseases (myelodysplasia), but their roles in allogeneic stem cell transplantation have not fully been studied in a large cohort of patients. We enrolled 797 patients who had been diagnosed with myelodysplasia at initial presentation and received transplantation via the Japan Marrow Donor Program. Targeted-capture sequencing was performed to identify mutations in 69 genes, together with copy-number alterations, whose effects on transplantation outcomes were investigated. We identified 1776 mutations and 927 abnormal copy segments among 617 patients (77.4%). In multivariate modeling using Cox proportional-hazards regression, genetic factors explained 30% of the total hazards for overall survival; clinical characteristics accounted for 70% of risk. TP53 and RAS-pathway mutations, together with complex karyotype (CK) as detected by conventional cytogenetics and/or sequencing-based analysis, negatively affected posttransplant survival independently of clinical factors. Regardless of disease subtype, TP53-mutated patients with CK were characterized by unique genetic features and associated with an extremely poor survival with frequent early relapse, whereas outcomes were substantially better in TP53-mutated patients without CK. By contrast, the effects of RAS-pathway mutations depended on disease subtype and were confined to myelodysplastic/myeloproliferative neoplasms (MDS/MPNs). Our results suggest that TP53 and RAS-pathway mutations predicted a dismal prognosis, when associated with CK and MDS/MPNs, respectively. However, for patients with mutated TP53 or CK alone, long-term survival could be obtained with transplantation. Clinical sequencing provides vital information for accurate prognostication in transplantation.
Introduction
Myelodysplastic syndromes (MDSs) and related myelodysplasias, including myelodysplastic/myeloproliferative neoplasms (MDS/MPNs), constitute a continuous spectrum of chronic myeloid neoplasms.1-4 Transitions between different disease subtypes are common and frequently terminate in secondary acute myeloid leukemia (sAML).2,5,6 Although allogeneic hematopoietic cell transplantation (HCT) remains the only curative treatment, patients with low-risk myelodysplasia can live for years without HCT. By contrast, high-risk diseases are associated with a profoundly truncated survival and consequently, for this group, the risk-benefit assessment of the utility of HCT balances in favor of recommending the treatment intervention, where recommendations for HCT have been based on clinical characteristics, such as those captured by the International Prognostic Scoring System (IPSS).7,8
Recently, revolutionized sequencing platforms have successfully been used to delineate a landscape of major genetic alterations involved in the pathogenesis of myelodysplasia.9-22 The impact of these alterations on disease phenotype and on prognosis has also been investigated in large-scale genotyping studies of untreated or minimally treated patients and, more recently, in the setting of HCT.16,23-27 However, these effects have not necessarily been fully explored in the latter context, for example, with respect to the influences of copy-number abnormalities,24 whose critical roles have been suggested by a recent study, albeit in a small cohort of transplanted patients.27 Moreover, transplantation outcomes are also affected by clinical factors, which also need to be incorporated into the analysis to clarify mutation-specific effects.28 Thus, whether molecular abnormalities should also be considered to determine the appropriateness of HCT has come into question.
In this study, we performed targeted-capture sequencing of pretransplant DNA samples from a large cohort of patients who had initially been diagnosed with myelodysplasia and received unrelated allogeneic HCT via the Japan Marrow Donor Program (JMDP), and investigated the effects of genetic alterations, including both gene mutations and copy-number abnormalities, on transplantation outcomes with relation to those of major clinical factors.
Methods
Patients’ characters and materials
A total of 797 patients with myelodysplasia who were subjected to HCT via JMDP at 136 centers and institutes were included in this study. The median duration from the time of initial diagnosis to the time of transplant was 9.4 months (range, 0.2-363 months), during which 110 (18%) of 609 patients with MDS (excluding refractory anemia with excess blasts [RAEB] in transformation [RAEB-t] in French-American-British [FAB] classification and myelodysplasia not otherwise specified) and MDS/MPN progressed to sAML in World Health Organization (WHO) classification. Disease type was re-evaluated at the time of transplantation in 760 patients (95%), of whom 283 (37%) were found to have higher-risk MDS (RAEB-1/2), 242 (32%) were found to have lower-risk disease (non-RAEB MDS), 46 (6%) were found to have MDS/MPN, and 189 (25%) were found to have sAML. Three hundred fourteen patients (39.4%) received HCT as their initial treatment of MDS and related diseases, whereas 104 patients (13.0%) and 122 patients (15.3%) were in complete or partial remission from prior therapies when they received HCT, respectively, and 231 patients (29.0%) had refractory/relapsed diseases. Stem cell source was bone marrow for 794 patients (99.6%) and mobilized peripheral blood for the remaining 3 (0.4%). Five hundred eleven (65.2%) patients received myeloablative-conditioning regimens, and a total body irradiation (TBI)-based regimen comprised 27.6% (220 cases). Other patient characteristics are detailed in Table 1. Clinical data were collected by the Japan Society for Hematopoietic Cell Transplantation (JSHCT) and the Japanese Data Center for Hematopoietic Cell Transplantation (JDCHCT) using the Transplant Registry Unified Management Program (TRUMP). Collected clinical data were processed with the Scripts for TRUMP data analyses.29,30 Genetic and clinical characteristics of relevant mutations and copy-number abnormalities were also investigated using an independent external cohort of 1577 patients with myelodysplasia, including 944 from MLL Munich Leukemia Laboratory25 and 633 from Cleveland Clinic.31 This study was approved by the institutional review board at the JMDP and Kyoto University.
Characteristics of 797 patients subjected to targeted-capture sequencing
| Characteristic . | No. of cases (%) . | Median (range) . |
|---|---|---|
| Patient sex (n = 797) | ||
| Male | 502 (63.0) | |
| Female | 295 (37.0) | |
| Age at diagnosis (n = 796), y | 51 (12-65) | |
| Age at transplantation (n = 797), y | 53 (16-66) | |
| Time from diagnosis to transplantation (n = 793), mo | 9.4 (0.2-363) | |
| Prior diagnosis (n = 797) | ||
| Lower-risk MDS | 276 (34.6) | |
| Higher-risk MDS | 307 (38.5) | |
| RAEB-t | 58 (7.3) | |
| MDS/MPN | 55 (6.9) | |
| Myelodysplasia not otherwise specified | 101 (12.7) | |
| IPSS (n = 532) | ||
| Low | 42 (7.9) | |
| Int-1 | 237 (44.5) | |
| Int-2 | 178 (33.5) | |
| High | 75 (14.1) | |
| Karyotype*(n = 770) | ||
| Low risk | 342 (44.4) | |
| Intermediate risk | 188 (24.4) | |
| High risk | 240 (31.2) | |
| Complex | 137 (17.8) | |
| Diagnosis at transplantation (n = 760) | ||
| Lower-risk MDS | 242 (30.4) | |
| Higher-risk MDS | 283 (35.5) | |
| sAML† | 189 (23.7) | |
| MDS/MPN | 46 (5.8) | |
| Disease status (n = 771) | ||
| No prior history of treatment | 314 (39.4) | |
| Complete remission | 104 (13.0) | |
| Partial remission | 122 (15.3) | |
| Refractory/relapsed disease | 231 (29.0) | |
| Year of transplantation (n = 794) | 2010 (1998-2014) | |
| Graft source (n = 797) | ||
| Bone marrow | 794 (99.6) | |
| Peripheral blood | 3 (0.4) | |
| Donor sex (n = 795) | ||
| Male | 555 (69.8) | |
| Female | 240 (30.2) | |
| HLA disparity (no. of serological mismatch) (n = 796) | ||
| 0/6 | 640 (80.4) | |
| 1/6 | 151 (19.0) | |
| 2/6 | 5 (0.6) | |
| Patient-donor sex disparity (n = 795) | ||
| No mismatch | 463 (58.2) | |
| Female-male | 193 (24.3) | |
| Male-female | 139 (17.5) | |
| GVHD prophylaxis (n = 797) | ||
| Tacrolimus and methotrexate | 606 (76.0) | |
| Cyclosporine and methotrexate | 129 (16.2) | |
| Others | 62 (7.8) | |
| Performance status at transplantation (n = 729) | ||
| 0-2 | 676 (92.7) | |
| ≥3 | 53 (7.3) | |
| History of RBC transfusion (n = 642) | ||
| + | 566 (88.2) | |
| − | 76 (11.8) | |
| History of platelet transfusion (n = 637) | ||
| + | 475 (74.6) | |
| − | 162 (25.4) | |
| Intensity of conditioning regimen (n = 784) | ||
| Myeloablative | 511 (65.2) | |
| Reduced-intensity | 273 (34.8) | |
| Conditioning regimen menu (n = 797) | ||
| TBI-based | 220 (27.6) | |
| HCT-CI score (n = 630) | ||
| 0 | 360 (57.1) | |
| 1-2 | 159 (25.2) | |
| ≥3 | 111 (17.6) |
| Characteristic . | No. of cases (%) . | Median (range) . |
|---|---|---|
| Patient sex (n = 797) | ||
| Male | 502 (63.0) | |
| Female | 295 (37.0) | |
| Age at diagnosis (n = 796), y | 51 (12-65) | |
| Age at transplantation (n = 797), y | 53 (16-66) | |
| Time from diagnosis to transplantation (n = 793), mo | 9.4 (0.2-363) | |
| Prior diagnosis (n = 797) | ||
| Lower-risk MDS | 276 (34.6) | |
| Higher-risk MDS | 307 (38.5) | |
| RAEB-t | 58 (7.3) | |
| MDS/MPN | 55 (6.9) | |
| Myelodysplasia not otherwise specified | 101 (12.7) | |
| IPSS (n = 532) | ||
| Low | 42 (7.9) | |
| Int-1 | 237 (44.5) | |
| Int-2 | 178 (33.5) | |
| High | 75 (14.1) | |
| Karyotype*(n = 770) | ||
| Low risk | 342 (44.4) | |
| Intermediate risk | 188 (24.4) | |
| High risk | 240 (31.2) | |
| Complex | 137 (17.8) | |
| Diagnosis at transplantation (n = 760) | ||
| Lower-risk MDS | 242 (30.4) | |
| Higher-risk MDS | 283 (35.5) | |
| sAML† | 189 (23.7) | |
| MDS/MPN | 46 (5.8) | |
| Disease status (n = 771) | ||
| No prior history of treatment | 314 (39.4) | |
| Complete remission | 104 (13.0) | |
| Partial remission | 122 (15.3) | |
| Refractory/relapsed disease | 231 (29.0) | |
| Year of transplantation (n = 794) | 2010 (1998-2014) | |
| Graft source (n = 797) | ||
| Bone marrow | 794 (99.6) | |
| Peripheral blood | 3 (0.4) | |
| Donor sex (n = 795) | ||
| Male | 555 (69.8) | |
| Female | 240 (30.2) | |
| HLA disparity (no. of serological mismatch) (n = 796) | ||
| 0/6 | 640 (80.4) | |
| 1/6 | 151 (19.0) | |
| 2/6 | 5 (0.6) | |
| Patient-donor sex disparity (n = 795) | ||
| No mismatch | 463 (58.2) | |
| Female-male | 193 (24.3) | |
| Male-female | 139 (17.5) | |
| GVHD prophylaxis (n = 797) | ||
| Tacrolimus and methotrexate | 606 (76.0) | |
| Cyclosporine and methotrexate | 129 (16.2) | |
| Others | 62 (7.8) | |
| Performance status at transplantation (n = 729) | ||
| 0-2 | 676 (92.7) | |
| ≥3 | 53 (7.3) | |
| History of RBC transfusion (n = 642) | ||
| + | 566 (88.2) | |
| − | 76 (11.8) | |
| History of platelet transfusion (n = 637) | ||
| + | 475 (74.6) | |
| − | 162 (25.4) | |
| Intensity of conditioning regimen (n = 784) | ||
| Myeloablative | 511 (65.2) | |
| Reduced-intensity | 273 (34.8) | |
| Conditioning regimen menu (n = 797) | ||
| TBI-based | 220 (27.6) | |
| HCT-CI score (n = 630) | ||
| 0 | 360 (57.1) | |
| 1-2 | 159 (25.2) | |
| ≥3 | 111 (17.6) |
In multivariate modeling, all of the missing data were imputed for each variable in all of the noninformative cases.
GVHD, graft-versus-host disease.
Karyotyping score is according to the criteria applied in IPSS.
sAML included patients with AML evolving from myelodysplasia, and those diagnosed with RAEB-t according to the French-American-British classification.
DNA sequencing
We performed targeted-capture sequencing as previously described (see “Methods” in the supplemental Appendix, available on the Blood Web site).21,25,32 RNA baits were designed for detection of oncogenic variants in 69 known driver genes implicated in myeloid malignancies (supplemental Table 1). A custom complementary RNA bait library to capture these 69 genes was generated (SureSelect; Agilent Technology). We designed the custom baits for capturing all coding exons of the 64 genes, hot-spot regions of 4 genes (NRAS: NM_002524 exon 2 [G12 and G13] and exon 3 [Q61]; HRAS: NM_005343 [G12 and G13] and exon 3 [Q61]; IDH1: NM_001282387 exon 4 [R132]; and IDH2: NM_002168 exon 4 [R140/R172]), and the promoter region of the TERT gene. Additional baits for 1158 single-nucleotide polymorphisms (SNPs) were also included to calculate genome-wide copy numbers (supplemental Table 2). These probes were deliberately selected so that they cover the human genome uniformly to allow for prospective detection of copy-number change and loss of heterozygosity (LOH) on the next-generation sequencing platform. Copy-number abnormalities33,34 were identified using the data of allele frequencies and sequenced depth of SNPs. This technique, referred to as CNACS, is under preparation for publication.35 DNA was extracted from the buffy coat of peripheral blood collected just before HCT. For 241 patients, amplified DNA was used for sequencing analysis. Methods of mutation calling21,25,32,36 and copy-number detection33,34 with calculation of their clone size32 were detailed in the supplemental Appendix. Complex karyotype (CK)–like abnormality was defined as ≥3 abnormal copy-number segments, including those with copy-number-neutral LOH or uniparental disomy (UPD) (“Methods” in the supplemental Appendix).
Statistical analyses
Statistical analyses were performed using R version 3.3.1. End points were overall survival, relapse-free survival, and nonrelapse mortality. Survival was calculated from the date of transplantation to the date of death from events under interest. Overall survival was evaluated using the Cox proportional hazards regression model. Relapse and nonrelapse mortality were analyzed with the Gray model assuming competing risks using R package “cmprsk” and “mstate.” Data were censored at the time patients were last known to be alive. Cases without achieving remission after transplant were assumed to have relapsed on day 0.1. We conducted multivariate analyses based on the Cox proportional hazards model, and optimal combination of covariates was selected with least absolute shrinkage and selection operator (lasso) using R package “glmnet” for overall survival, and with stepwise regression for relapse-free survival and nonrelapse mortality. We performed multiple imputation toward missing values with the bootstrap-based expectation-maximization method using R package “Amelia” and created 100 imputed complete data sets. Objective variables (survival and relapse data) were not imputed. We performed survival analysis to all 100 data sets separately and combined the obtained results by the Rubin's rule37 with R package “cat.” All P values were calculated with the use of 2-sided tests. Multiple testing was corrected with Benjamini-Hochberg q value. Details of statistical analyses are provided in “Methods” in the supplemental Appendix.
Results
Patients
We included a total of 797 patients who had been diagnosed with MDS or MDS/MPN at initial presentation and received unrelated HCT through the JMDP between 1998 and 2014 at 16 years of age or older (median, 53 years; range, 16-66 years), and from whom peripheral blood-derived DNA had been isolated just before HCT and archived for sequencing (Table 1; supplemental Figure 1). Written informed consent was obtained from all participants. Karyotyping data were available in 770 patients.
Detection of driver alterations
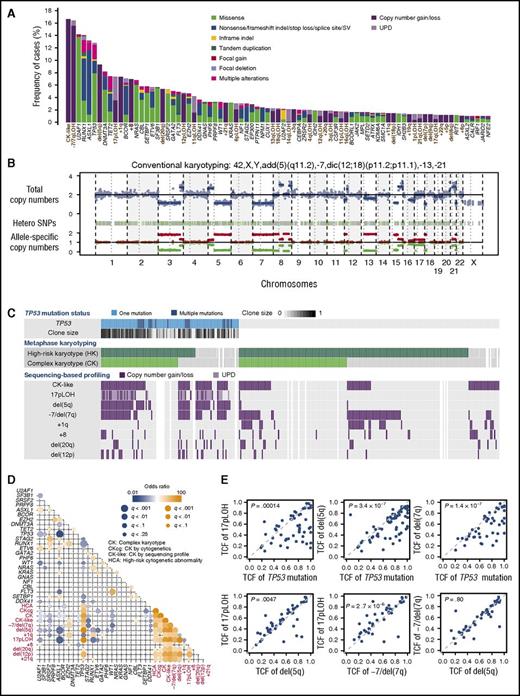
We performed targeted-capture sequencing using a custom bait library which included 69 known driver genes implicated in myeloid malignancies and an additional 1158 SNPs to calculate genome-wide copy numbers (supplemental Tables 1 and 2). With a mean coverage of 888× (supplemental Figure 2; supplemental Table 3), targeted-capture sequencing identified 1776 oncogenic driver mutations in 597 of 797 patients. Mutations involved 68 genes and consisted of 1033 nonsynonymous single-nucleotide variations and 369 insertions/deletions (indels), as well as 115 structural variations (Figure 1A; supplemental Table 4). Copy-number profiling was reliably performed in 763 samples (95.7%), in which 927 abnormal copy-number segments in 318 samples were detected (supplemental Figure 3). In addition to arm level copy-number gains and losses, sequencing-based copy-number profiling detected copy-neutral events (UPDs) and submicroscopic lesions that would not have come under detection with conventional cytogenetics (Figure 1B; supplemental Figure 4).
Summary of targeted-capture sequencing in 797 patients. (A) Frequency distribution of 81 genetic abnormalities, including 48 mutations and 32 abnormal chromosomal segments, which are designated along the bottom. Abnormal chromosomal segments are indicated in brown. Type of mutation and chromosomal lesions are also indicated. (B) A representative copy-number profile obtained from sequencing-based analysis, showing multiple alterations in chromosomal copy numbers, conforming to conventional cytogenetics. (C) Summary of major chromosomal copy-number abnormalities in 295 patients with mutated TP53, CK, and high-risk copy-number lesions (≥3 abnormalities or chromosome 7 abnormality, according to IPSS scoring system) as detected by either conventional cytogenetics and/or sequencing-based copy-number profiling. TP53-mutated tumor cell fractions are depicted in color gradient. (D) Correlation between mutations and copy-number abnormalities found in >3% in the entire cohort. Correlation coefficients and associated q values are indicated by the size of circles and color gradient. (E) Diagonal plots from various combinations of TCFs carrying TP53 mutations, del(5q), 17pLOH, and −7/del(7q), detected by sequencing-based copy-number profiling.
Summary of targeted-capture sequencing in 797 patients. (A) Frequency distribution of 81 genetic abnormalities, including 48 mutations and 32 abnormal chromosomal segments, which are designated along the bottom. Abnormal chromosomal segments are indicated in brown. Type of mutation and chromosomal lesions are also indicated. (B) A representative copy-number profile obtained from sequencing-based analysis, showing multiple alterations in chromosomal copy numbers, conforming to conventional cytogenetics. (C) Summary of major chromosomal copy-number abnormalities in 295 patients with mutated TP53, CK, and high-risk copy-number lesions (≥3 abnormalities or chromosome 7 abnormality, according to IPSS scoring system) as detected by either conventional cytogenetics and/or sequencing-based copy-number profiling. TP53-mutated tumor cell fractions are depicted in color gradient. (D) Correlation between mutations and copy-number abnormalities found in >3% in the entire cohort. Correlation coefficients and associated q values are indicated by the size of circles and color gradient. (E) Diagonal plots from various combinations of TCFs carrying TP53 mutations, del(5q), 17pLOH, and −7/del(7q), detected by sequencing-based copy-number profiling.
Combining mutations and copy-number abnormalities, a total of 2703 genetic alterations were detected in 617 samples with a median of 4 per sample (range, 1-21), of which 500 samples had multiple alterations (supplemental Figures 5 and 6). On average, the highest number of alterations was found in sAML, followed by MDS/MPNs, higher-risk and lower-risk MDS (P = 1.1 × 10−15) (supplemental Figure 7). The frequency distribution of genetic alterations in this cohort differed substantially from what has been reported in untreated or only minimally treated myelodysplasia patients.25,26 There was an underrepresentation of SF3B1, TET2, and SRSF2 mutations, whereas copy-number abnormalities including −7/del(7q), del(5q), and 17pLOH, as well as mutations in TP53, U2AF1, NRAS, BCOR, and FLT3 (Figure 1A), were overrepresented, likely due to a higher proportion of high-risk patients.
Correlations between genetic abnormalities
Strong positive and negative correlations, that is, trends of co-occurrence or mutual exclusiveness, respectively, were observed between several genetic alterations (Figure 1D). Most conspicuous among these were those found between TP53 mutations and associated abnormalities. TP53 mutations showed significant positive correlations with a number of copy-number abnormalities, including 17pLOH, del(5q), −7/del(7q), del(12p), del(20q), and +21q, which in turn were in close association with each other, participating in the generation of CK-like abnormality. Significant negative correlations were also observed between TP53 mutations and other mutations, including those affecting ASXL1, RUNX1, U2AF1, SF3B1, and STAG2, likely reflecting a generally lower mutational burden (1.44 per sample) in TP53-mutated patients, compared with TP53-unmutated ones (2.16 per sample; P = .00028) (supplemental Figure 5). These positive and negative correlations were largely recapitulated in an external cohort of 1577 cases with different subtypes of myelodysplasia in nontransplantation settings (supplemental Figure 8; supplemental Table 5).
CK-like abnormality was detected in 62 of 102 samples with mutated TP53 in the JMDP cohort, which were mostly accompanied by del(5q) (n = 58) and less frequently by 17pLOH (n = 48) (Figure 1C). Almost always affecting the TP53 locus, the 17pLOH was associated with homozygous or hemizygous TP53 mutations and, together with multiple TP53 mutations found in an additional 12 samples, was involved in biallelic TP53 inactivation. Of the remaining 40 TP53-mutated cases, 24 had been reported to have CK in conventional cytogenetics, even though no CK-like abnormality was detected by sequencing-based analysis, which was successfully performed on 17 of the 24 samples. The exact reasons for this apparent discrepancy were unknown, but most likely to be explained by different cell sources, sampling time, and target cell population, as well as the difference in sensitivity between assays.38-40 Thus, taking this into consideration, most of the TP53-mutated cases had a CK or a CK-like abnormality, together with accompanying 17pLOH and del(5q). −7/del(7q) has also been closely associated with a CK, which was also confirmed by sequencing-based copy-number profiling (n = 47). Of interest, the TP53 mutation showed a larger tumor cell fraction (TCF) than 17pLOH, del(5q), and −7/del(7q). By contrast, TCFs for the latter 3 lesions were largely similar in most cases, although a small but significant difference in TCF was detected, when comparisons were made between 17pLOH, del(5q), and −7/del(7q) (Figure 1E). Thus, in the majority of cases, the dominant clones were likely to originate from TP53-mutated founding clones, which subsequently acquired 17pLOH, followed by del(5q) and −7/del(7q).
Effects of genetic alterations on transplantation outcomes
Effects of genetic alterations on transplantation outcomes first became evident from a significant association of the number of genetic lesions with both short-term (at day 100) and long-term survival following HCT, relapse rate, and long-term nonrelapse mortality, which were seen both for the total number of lesions and for the numbers of both mutations and copy-number abnormalities (Figure 2A; supplemental Table 6). In univariate analysis, we identified a total of 4 mutations and 9 copy-number lesions, together with 11 clinical factors, that were found in >5% of all patients and significantly associated with posttransplant survival (Figure 2B; supplemental Table 7). Although previous studies identified mutations in DNMT3A, TET2, RUNX1, and ASXL1 as being significantly associated with shorter survival,24,27 these were not confirmed in the present cohort, except for a modest effect of RUNX1 mutations (Figure 2B; supplemental Figure 9). No significant effects of conditioning regimens on outcomes were observed in the cohort or in any disease/mutation subtypes (supplemental Table 7; supplemental Figure 10).
Effects of gene mutations on overall survival after stem cell transplantation. (A) Overall survival and incidence of relapse in patients carrying different numbers of genetic abnormalities, which are shown in indicated colors. Hazard ratios according to numbers of genetic events are provided. (B) Volcano plot of hazard ratios (in horizontal axis) and corresponding P values (in vertical axis) according to univariate analysis of the effect of individual genetic and clinical factors on overall survival. Size of circles corresponds to the fraction of patients carrying indicated factors, among which significant (P < .05) ones are annotated. Clinical, cytogenetic, and genetic factors are shown in green, blue, and red, respectively. (C) Relative contribution of clinical and genetic factors to the log hazard of overall survival in Cox proportional hazards regression modeling. Contribution of both combined and individual factors are depicted in horizontal and vertical axis, respectively. (D) Effects of the number of genetic factors, as indicated by color, on overall survival are shown in Kaplan-Meier curves. Hazard ratios relative to zero genetic factor are presented. One case with 4 genetic factors was incorporated in cases with 3. CKcg, CK by conventional cytogenetics; CK-like, CK by sequencing analysis; Dx, diagnosis; HR, hazard ratio; HSCT, hematopoietic stem cell transplantation; PLT, platelet; PS, performance status; Ref/Rel, refractory or relapse.
Effects of gene mutations on overall survival after stem cell transplantation. (A) Overall survival and incidence of relapse in patients carrying different numbers of genetic abnormalities, which are shown in indicated colors. Hazard ratios according to numbers of genetic events are provided. (B) Volcano plot of hazard ratios (in horizontal axis) and corresponding P values (in vertical axis) according to univariate analysis of the effect of individual genetic and clinical factors on overall survival. Size of circles corresponds to the fraction of patients carrying indicated factors, among which significant (P < .05) ones are annotated. Clinical, cytogenetic, and genetic factors are shown in green, blue, and red, respectively. (C) Relative contribution of clinical and genetic factors to the log hazard of overall survival in Cox proportional hazards regression modeling. Contribution of both combined and individual factors are depicted in horizontal and vertical axis, respectively. (D) Effects of the number of genetic factors, as indicated by color, on overall survival are shown in Kaplan-Meier curves. Hazard ratios relative to zero genetic factor are presented. One case with 4 genetic factors was incorporated in cases with 3. CKcg, CK by conventional cytogenetics; CK-like, CK by sequencing analysis; Dx, diagnosis; HR, hazard ratio; HSCT, hematopoietic stem cell transplantation; PLT, platelet; PS, performance status; Ref/Rel, refractory or relapse.
Next, to evaluate the relative effects of clinical and genetic factors on posttransplant survival, we performed Cox proportional hazards regression modeling, using 22 variables significantly associated with overall survival in univariate analysis. All patients with any evidence of CK from conventional cytogenetics and/or sequencing-based analysis were defined as having CK. After 100 rounds of multiple imputation of missing data using a bootstrap-based expectation-maximization method, followed by modeling on the basis of lasso, 14 variables were stably incorporated in all imputations, of which 12 were shown to significantly affect the model, including 4 genetic lesions: mutations in NRAS, TP53, and CBL as well as CK and 8 clinical variables, performance status (≥3), age at HCT (≥41.5), hematopoietic stem cell transplantation–specific comorbidity index (HCT-CI) (score ≥3), female donor to male, diagnosis (sAML) and disease status (complete remission [CR]) at transplant, time from initial diagnosis to transplant, and red blood cell (RBC) transfusion history (Table 2; supplemental Figure 11).
Variables stably incorporated in multivariate model for posttransplant survival
| Covariates . | n (%) . | HR (95% CI) . | Coeff. . | P . |
|---|---|---|---|---|
| Genetic factors | ||||
| NRAS mutation | 54 (6.8) | 1.64 (1.14-2.35) | 0.49 | .0075 |
| TP53 mutation | 101 (12.7) | 1.49 (1.10-2.00) | 0.40 | .0096 |
| CBL mutation | 46 (5.8) | 1.55 (1.06-2.26) | 0.44 | .024 |
| CK | 218 (27.4) | 1.45 (1.01-2.09) | 0.37 | .046 |
| High-risk cytogenetic abnormality | 303 (38.1) | 1.29 (0.92-1.82) | 0.26 | .14 |
| Clinical factors | ||||
| Age at HCT, ≥41.5 y | 601 (75.8) | 1.69 (1.29-2.22) | 0.53 | .00014 |
| PS, ≥3 | 63 (8.2) | 1.90 (1.36-2.68) | 0.64 | .00021 |
| HCT-CI, score ≥3 | 143 (18.0) | 1.52 (1.17-1.98) | 0.42 | .0016 |
| RBC transfusion history, negative | 99 (12.7) | 0.52 (0.34-0.81) | −0.65 | .0038 |
| Female donor to male | 139 (17.6) | 1.43 (1.12-1.84) | 0.36 | .0042 |
| Dx at HCT: sAML | 203 (25.6) | 1.32 (1.04-1.68) | 0.28 | .020 |
| Days from Dx to HCT, ≥1275 d | 105 (13.3) | 0.64 (0.44-0.94) | −0.44 | .022 |
| Disease status | ||||
| CR | 106 (13.7) | 0.69 (0.48-0.99) | −0.37 | .044 |
| Ref/Rel | 237 (29.7) | 1.23 (0.97-1.55) | 0.20 | .086 |
| Covariates . | n (%) . | HR (95% CI) . | Coeff. . | P . |
|---|---|---|---|---|
| Genetic factors | ||||
| NRAS mutation | 54 (6.8) | 1.64 (1.14-2.35) | 0.49 | .0075 |
| TP53 mutation | 101 (12.7) | 1.49 (1.10-2.00) | 0.40 | .0096 |
| CBL mutation | 46 (5.8) | 1.55 (1.06-2.26) | 0.44 | .024 |
| CK | 218 (27.4) | 1.45 (1.01-2.09) | 0.37 | .046 |
| High-risk cytogenetic abnormality | 303 (38.1) | 1.29 (0.92-1.82) | 0.26 | .14 |
| Clinical factors | ||||
| Age at HCT, ≥41.5 y | 601 (75.8) | 1.69 (1.29-2.22) | 0.53 | .00014 |
| PS, ≥3 | 63 (8.2) | 1.90 (1.36-2.68) | 0.64 | .00021 |
| HCT-CI, score ≥3 | 143 (18.0) | 1.52 (1.17-1.98) | 0.42 | .0016 |
| RBC transfusion history, negative | 99 (12.7) | 0.52 (0.34-0.81) | −0.65 | .0038 |
| Female donor to male | 139 (17.6) | 1.43 (1.12-1.84) | 0.36 | .0042 |
| Dx at HCT: sAML | 203 (25.6) | 1.32 (1.04-1.68) | 0.28 | .020 |
| Days from Dx to HCT, ≥1275 d | 105 (13.3) | 0.64 (0.44-0.94) | −0.44 | .022 |
| Disease status | ||||
| CR | 106 (13.7) | 0.69 (0.48-0.99) | −0.37 | .044 |
| Ref/Rel | 237 (29.7) | 1.23 (0.97-1.55) | 0.20 | .086 |
Multivariate analysis was conducted for 793 patients whose outcome data were available.
95% CI, 95% confidence interval; Coeff., coefficient; CR, complete remission; Dx, diagnosis; HR, hazard ratio; PS, performance status; Ref/Rel, refractory or relapse.
Modeling was also performed 100 times using 80% samples and validated in the remaining 20% using c-statistics, where the identical set of variables was stably selected in ≥95 of 100 validations with a mean c-statistic score of 0.68 (supplemental Figure 12). TP53 and CBL mutations, and NRAS mutation, were also selected as significant factors in multivariate modeling of relapse and nonrelapse death, respectively. High-risk cytogenetic abnormality (ie, CK and/or any abnormality of chromosome 7) was also associated with nonrelapse death (Figure 3; supplemental Methods; supplemental Figure 13).
Multivariate analyses for relapse-free survival and nonrelapse mortality. Forest plots showing the results of multivariate analysis for relapse-free survival (A) and nonrelapse mortality (B). The covariates selected in the final model are shown. Positively and negatively significant factors are shown in red and blue, respectively. Missing data were complemented by multiple imputation and the final model was constructed with competing regression model using covariates which were stably selected in all the 100 imputed data sets.
Multivariate analyses for relapse-free survival and nonrelapse mortality. Forest plots showing the results of multivariate analysis for relapse-free survival (A) and nonrelapse mortality (B). The covariates selected in the final model are shown. Positively and negatively significant factors are shown in red and blue, respectively. Missing data were complemented by multiple imputation and the final model was constructed with competing regression model using covariates which were stably selected in all the 100 imputed data sets.
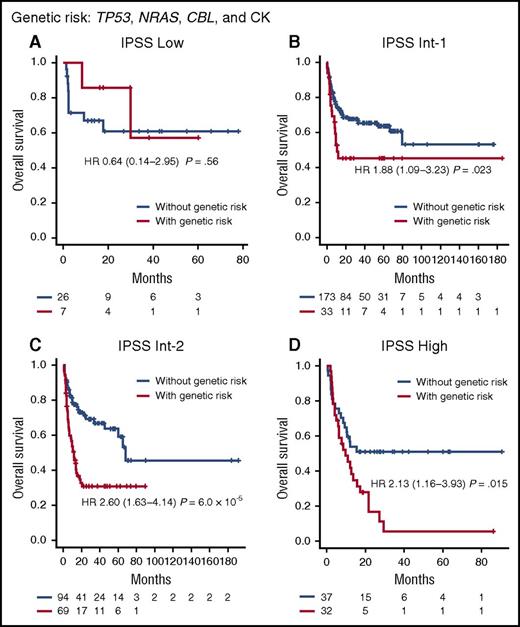
In this particular multivariate model for overall survival, clinical characteristics of patients before HCT were the predominant determinants of posttransplant outcomes, explaining as much as 70% of the total hazard overall. Nevertheless, genetic factors still influence HCT outcomes substantially, accounting for 30% of the total hazard, where copy-number alterations were most influential (70% of the hazard from genetic alterations), compared with mutations in TP53, CBL, and NRAS (Figure 2C; supplemental Figure 14). Patients were stratified into distinct risk subgroups showing substantially different survival according to the number of genetic factors associated with a given patient (Figure 2D). The independent effects of genetic factors (TP53, CBL, and NRAS mutations and CK), distinct from clinical variables, were also tested by assessing their effect on each IPSS subgroup and disease subtype with respect to prediction of survival; patients carrying 1 or more of the above 4 genetic lesions (their combination is shown in supplemental Figure 15) had significantly inferior posttransplant survival within each IPSS subgroup (Figure 4; supplemental Figure 16) and each disease subtype (supplemental Figure 17), suggesting that the relevant genetic factors confer independent risks from those variables included in the IPSS.41
Effects of genetic risk on survival within IPSS-defined risk groups. Kaplan-Meier curves for patients with (red) and without (blue) genetic risk are depicted for each IPSS-defined risk group: (A) low; (B) Int-1; (C) Int-2; (D) high. Cases whose initial diagnosis was MDS/MPN were excluded from the analysis. Genetic risk factors included TP53, NRAS, and CBL mutations and CK. Patients carrying 1 or more of the 4 genetic lesions were defined as cases with genetic risk.
Effects of genetic risk on survival within IPSS-defined risk groups. Kaplan-Meier curves for patients with (red) and without (blue) genetic risk are depicted for each IPSS-defined risk group: (A) low; (B) Int-1; (C) Int-2; (D) high. Cases whose initial diagnosis was MDS/MPN were excluded from the analysis. Genetic risk factors included TP53, NRAS, and CBL mutations and CK. Patients carrying 1 or more of the 4 genetic lesions were defined as cases with genetic risk.
Effects of TP53 mutations and CK
Associated with independent risks for shorter survival irrespective of disease subtypes (supplemental Figure 18), both TP53 mutations and CK define a set of subgroups showing substantial overlaps between each other (Figure 1C).16,42-44 The overlaps were most prominent within the TP53-mutated patients; 86 of 98 cases with mutated TP53 (88%) had a CK, whereas only 42% of cases with a CK had mutated TP53 (supplemental Table 8; supplemental Figure 19). This was in contrast to a much higher TP53 mutation rate in patients with CK in the nontransplantation cohort (72%), which is partly explained not only by the fact that the highly aggressive clinical course in these patients precluded survival to transplantation, but also by the fact that without sequencing-based analysis, some patients with CK had not been detected. Regardless of prior therapy, disease subtype, conditioning regimen, performance status, and organ dysfunction prior to transplant, the outcome of patients with mutated TP53 and CK undergoing HCT was poor (Figure 5A; supplemental Figures 20 and 21; supplemental Table 8). The median survival for patients with both lesions was only 4.8 months; 38% of patients died prior to day 100 and >80% within 2 years after transplantation. The leading cause of death was early relapse, which explained ∼60% of the cases (Figure 3; supplemental Figure 13). The fraction of TP53-mutated cells significantly correlated with the shorter time from transplantation to death (P = .00052) and worse overall survival (P = .0019),45 although the effect was smaller than that of CK (supplemental Figure 22). The very poor clinical outcome for this combination of abnormalities was also seen in the external, nontransplantation cohort (Figure 5C).
Effects of TP53 and CK on survival and relapse. (A-B) Effects of TP53 mutation and CK as determined by conventional cytogenetics (CKcg), sequencing-based analysis (CK-like), and combined assays as indicated by color. (C) Effects of TP53 mutation and CK on overall survival in the external cohort. Overall survivals (A,C) and cumulative incidence of relapse (B) for the patients with indicated abnormalities are shown in each panel. The numbers of patients at risk are shown at indicated time points along the bottom (A,C). Hazard ratios are also indicated in corresponding colors with 95% confidence intervals.
Effects of TP53 and CK on survival and relapse. (A-B) Effects of TP53 mutation and CK as determined by conventional cytogenetics (CKcg), sequencing-based analysis (CK-like), and combined assays as indicated by color. (C) Effects of TP53 mutation and CK on overall survival in the external cohort. Overall survivals (A,C) and cumulative incidence of relapse (B) for the patients with indicated abnormalities are shown in each panel. The numbers of patients at risk are shown at indicated time points along the bottom (A,C). Hazard ratios are also indicated in corresponding colors with 95% confidence intervals.
In contrast to the dismal prognosis of TP53-mutated patients with CK, patients having TP53 mutations alone (12% of TP53-mutated patients) had a significantly better survival posttransplant (73% alive at 60 months [95% confidence interval, 51%-100%]) (Figure 5A-B; supplemental Table 9). These patients were extracted only after rigorously excluding an additional 29 patients having “masked” CK using sequencing-based copy-number analysis (supplemental Figure 19). Similarly, TP53 mutation-alone patients had a significantly better overall survival in the external nontransplantation cohort of myelodysplasia, compared with those with both TP53 and CK (P = 3.7 × 10−7) (Figure 5C). For CK-alone patients, the posttransplant survival was worse than patients with TP53 mutation-alone (P = .049) or those without TP53 or CK (P = 4.3 × 10−8), but significantly better than that of patients having TP53 mutation and CK (P = .0020) (Figure 5A). Similar results were also obtained in nontransplant settings (Figure 5C).
RAS-pathway mutations
RAS signaling is one of the major pathways commonly affected by somatic mutations in myeloid neoplasms,11,25,26,46 where NRAS and CBL represent the 2 most frequently affected genes, mutated in 54 patients (6.8%) and 46 patients (5.8%) in our cohort, respectively. Combined with less frequent mutations in KRAS, NF1, and PTPN11, RAS-pathway mutations accounted for 135 (17%) of 797 patients. Distinct RAS-pathway mutations yielded similar overall survival results (Figure 6A), and in multivariate modeling with all 5 genes combined, RAS-pathway mutations were significantly associated with worse overall survival, together with TP53 mutations and CK (supplemental Figure 23). Unlike the paucity of mutations in TP53-mutated patients, RAS-pathway mutated patients were characterized by a high mutational burden, with no significant difference among different RAS-pathway mutations (supplemental Tables 10 and 11), although the effects of RAS-pathway mutations on survival were independent of the number of mutations (supplemental Figure 24).47,48
Effects of RAS-pathway mutations on overall survival. (A) Effects of combined RAS-pathway mutations on overall survival. Kaplan-Meier curves for individual RAS-pathway mutations, including NRAS, KRAS, CBL, NF1, and PTPN11 genes are also presented in indicated colors. (B) Effects of RAS-pathway mutations depending on disease subtype. (C) Effects of RAS-pathway mutations for sAML subtype depending on initial diagnosis. Kaplan-Meier curves for disease subtypes (B) and initial diagnosis (C) are presented. The numbers of patients at risk are shown at indicated time points along the bottom (A-C). Hazard ratios are also indicated in corresponding colors with 95% confidence intervals.
Effects of RAS-pathway mutations on overall survival. (A) Effects of combined RAS-pathway mutations on overall survival. Kaplan-Meier curves for individual RAS-pathway mutations, including NRAS, KRAS, CBL, NF1, and PTPN11 genes are also presented in indicated colors. (B) Effects of RAS-pathway mutations depending on disease subtype. (C) Effects of RAS-pathway mutations for sAML subtype depending on initial diagnosis. Kaplan-Meier curves for disease subtypes (B) and initial diagnosis (C) are presented. The numbers of patients at risk are shown at indicated time points along the bottom (A-C). Hazard ratios are also indicated in corresponding colors with 95% confidence intervals.
In contrast to TP53 mutations and CK, which were strongly associated with shorter survival regardless of disease subtype (supplemental Figures 20 and 21), the effects of RAS-pathway mutations depended on disease subtype (Figure 6B); RAS-pathway mutations negatively affected posttransplant survival among patients with sAML and MDS/MPNs, but not among MDS patients. In addition, the negative impact of RAS-pathway mutations in sAML was only seen among patients who had been diagnosed with RAEB-t at the initial presentation, but not among those who were progressed from MDS (Figure 6C). This was expected, because these RAEB-t patients should have been better diagnosed with primary AML, according to the WHO classification, and as such, are generally expected to have a substantially better prognosis than true sAML patients, except for some borderline cases, such as RAS-pathway mutated patients, who were more likely to represent bona fide MDS-derived AML.31,49-54 Taken together, the negative effects of RAS-pathway mutations appeared to be confined to MDS/MPN and RAEB-t (primary AML in the WHO classification) patients, but not those with MDS and sAML progressed from WHO-defined MDS.
Discussion
It has long been recognized that survival in MDS is associated with straightforward clinical variables, and these have been used to determine appropriate initial and subsequent therapy. More recently, discrete molecular abnormalities have been identified in >90% of patients, and have also been applied therapeutically and for prognostication. Understanding the interplay between clinical and molecular determinants in making HCT recommendations is thus critical.
We performed a large-scale, high-throughput sequencing analysis using archived DNA from patients with different MDS subtypes, MDS/MPNs, and sAML who were transplanted via the JMDP. Together with similar findings from other reports,24,27 our results confirm the significant effects of genetic abnormalities on the outcome of HCT for patients with these myeloid malignancies. In addition, we show that independent of clinical factors, genetic abnormalities, including TP53 and RAS-pathway mutations as well as CK and other high-risk cytogenetic abnormalities, explain as much as 30% of the total hazard for survival associated with HCT in myelodysplasia, meaning that 70% is still associated with clinical factors. The risk of posttransplant death for individual patients is calculated based on the multivariate model (supplemental Table 12).
The strong adverse effect of TP53 mutations in combination with CK, previously suggested in a small cohort of patients,27 has convincingly been demonstrated in the current study using a larger number of patients. TP53-mutated cases associated with CK represent a distinct subset of myeloid neoplasms, irrespective of clinical/pathological diagnosis, and are unlikely to benefit from most therapies, including transplantation. For the accurate diagnosis of these patients, detection of a masked CK in TP53-mutated patients is critical, as these patients had as poor a survival with HCT as those whose CK was detected through conventional cytogenetics, underscoring the role of sequencing-base analysis. By contrast, in the absence of CK, patients with TP53 mutations had excellent survival with HCT, although this needs to be confirmed by an independent cohort. It is also important to precisely evaluate the outcome of CK-negative TP53-mutated patients in a nontransplantation setting.
The disease subtype-specific effects of RAS-pathway mutations in patients undergoing HCT were unexpected. These mutations were relatively late events in MDS, and are implicated in the progression to sAML, whereas they represent earlier events in MDS/MPNs.11,47 The poor outcome of RAS-pathway mutated patients, especially those with MDS/MPNs, did not appear to be overcome by conditioning regimen (supplemental Figure 25). Thus, novel strategies for transplantation for these patients should be investigated, such as the use of RAS inhibitors.55,56
Effects of genetic alterations in HCT for MDS were investigated in 2 previous studies.24,27 Bejar et al performed targeted sequencing of 40 genes in 87 MDS patients who received HCT and identified TP53, TET2, and DNMT3A mutations as significant risk factors for survival after HCT.27 More recently, Della Porta et al analyzed 401 MDS or sAML patients with HCT for mutations in 34 genes and concluded that mutations in ASXL1, RUNX1, and TP53 were independent risk factors for survival.24 Throughout the 3 studies including ours, strong negative effects of TP53 mutations and CK are reproducible findings, whereas no unanimous results were obtained for other mutations. We speculate that the discrepant results for the latter abnormalities might be due to differences in the ethnicity, disease subtype, cohort size, and/or variables incorporated into multivariate analysis. Difference in the platform for copy-number detection, that is, cytogenetics vs combined cytogenetics and sequencing-based assay, might have also affected the results. For example, in our cohort, negative effects of RAS-pathway mutations were only detected in the MDS/MPN subtype, which was not included in the previous studies. A meta-analysis combining these different studies would be a possible approach to address these discrepant results.
In conclusion, MDS, MDS/MPN, and sAML patients with mutations in TP53 or RAS-pathway or CK have poor post-HCT outcomes. Patients with both TP53 mutations and CK have particularly poor survival, as do those with both MDS/MPN and RAS-pathway mutations, and consideration for therapies other than HCT should be made for these patients, whereas those with TP53 mutations without CK do comparatively well with HCT. Recommendations for HCT should be based on both clinical and molecular factors.
The sequencing data reported in this article have been deposited in the European Genome-Phenome Archive (accession number EGAS00001001949).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors appreciate all of the staff of the JMDP for their assistance and all physicians and nursing staff of all transplantation teams in Japan for providing excellent patient care and reporting transplant data. The authors also express their deep gratitude to those patients who have provided consent to use their biological materials and clinical data, and to the volunteers who have donated stem cells to the patients.
This work was supported by Grant-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science (23249052, 22134006, and 21790907 [Kyoto; S.O.], 26115009 [Nagoya; S.M.], and 16H05338 [Kyoto; H.M.]), the Tailor-made Medical Treatment Program from Japan Agency for Medical Research and Development (15km0305018h0101 [Kyoto; H.M.]), the Project for Cancer Research and Therapeutics Evolution (P-CREATE) from Japan Agency for Medical Research and Development (16cm0106501h0001 [Kyoto; S.O.] and 16cm0106422h0001 [Kyoto; T.Y.]), the Foundation for the National Institutes of Health (Bethesda, MD) (National Heart, Lung, and Blood Institute RO1HL-082983, National Institute of General Medical Sciences U54 RR019391, and National Heart, Lung, and Blood Institute K24 HL-077522 [Cleveland, OH; J.P.M.]), the Scott Hamilton CARES grant (Cleveland, OH; H.M.), and the Edward P. Evans Foundation (Cleveland, OH; M.A.S.).
Authorship
Contribution: T.Y., Y.A., K.H., and S.O. designed the study; Y.A., M.O., C.H., W.K., H.I., Y.K., M.A.S., J.P.M., T.H., and Y.M. collected clinical data and DNA specimens; T.Y., Y. Shiozawa, Y.I.-Y., K.Y., H.S., Y. Nagata, K.K., K.H., and M.S. performed sample preparation and sequencing; T.Y., Y. Nannya, Y. Shiozawa, K.Y., Y. Shiraishi, H.S., Y. Nagata, K.K., Y. Sato, N.K., K.C., H.T., H.U., M.M.N., B.P., C.H., W.K., Y.K., S.M., and H.M. analyzed data; T.Y., Y. Nannya, K.A., and K.M. performed statistical analysis; T.Y., Y. Nannya, M.A.S., H.M., and S.O. wrote the manuscript; and all of the authors reviewed and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Seishi Ogawa, Pathology and Tumor Biology, Graduate School of Medicine, Kyoto University, Yoshida-Konoe-cho, Sakyo-ku, Kyoto, 606-8501, Japan; e-mail: sogawa-tky@umin.ac.jp.
References
Author notes
T.Y., Y. Nannya, Y.A., and Y. Shiozawa contributed equally to this study.