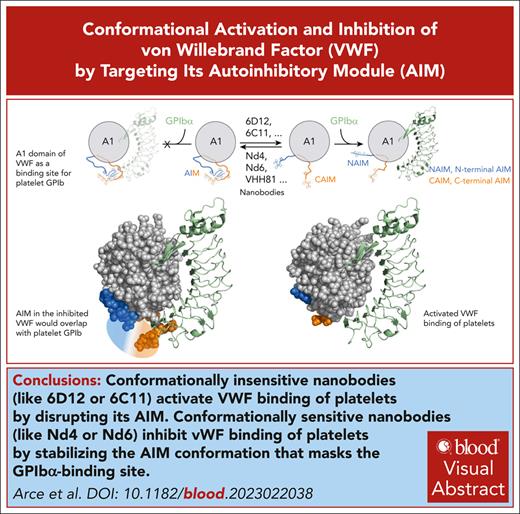
Conformationally insensitive nanobodies activate VWF binding of platelets by disrupting its NAIM.
Conformationally sensitive nanobodies inhibit VWF binding of platelets by stabilizing the AIM conformation that masks the GPIbα-binding site.
Visual Abstract
Activation of von Willebrand factor (VWF) is a tightly controlled process governed primarily by local elements around its A1 domain. Recent studies suggest that the O-glycosylated sequences flanking the A1 domain constitute a discontinuous and force-sensitive autoinhibitory module (AIM), although its extent and conformation remains controversial. Here, we used a targeted screening strategy to identify 2 groups of nanobodies. One group, represented by clone 6D12, is conformation insensitive and binds the N-terminal AIM (NAIM) sequence that is distal from A1; 6D12 activates human VWF and induces aggregation of platelet-rich plasma at submicromolar concentrations. The other group, represented by clones Nd4 and Nd6, is conformation sensitive and targets the C-terminal AIM (CAIM). Nd4 and Nd6 inhibit ristocetin-induced platelet aggregation and reduce VWF-mediated platelet adhesion under flow. A crystal structure of Nd6 in complex with AIM-A1 shows a novel conformation of both CAIM and NAIM that are primed to interact, providing a model of steric hindrance stabilized by the AIM as the mechanism for regulating GPIbα binding to VWF. Hydrogen-deuterium exchange mass spectrometry analysis shows that binding of 6D12 induces the exposure of the GPIbα-binding site in the A1 domain, but binding of inhibitory nanobodies reduces it. Overall, these results suggest that the distal portion of NAIM is involved in specific interactions with CAIM, and binding of nanobodies to the AIM could either disrupt its conformation to activate VWF or stabilize its conformation to upkeep VWF autoinhibition. These reported nanobodies could facilitate future studies of VWF functions and related pathologies.
Introduction
von Willebrand factor (VWF) is a multimeric plasma glycoprotein essential for primary hemostasis.1 VWF can activate and capture platelets to initiate thrombus formation by sensing and responding to altered blood flow.2 Once activated, the A1 domain of VWF becomes exposed and binds to the ligand-binding domain (LBD) of glycoprotein (GP)Ibα on the platelet surface. Circulating VWF multimers do not bind platelets, indicating that VWF is autoinhibited under normal flow conditions. The whereabouts of the autoinhibitory elements in VWF and their molecular nature have been under investigation. Contrary to prior beliefs, multimeric VWF in solution may not adopt a collapsed globular shape, and a critical force threshold for uncoiling the multimer may not exist.3,4 Because global extension of a VWF multimer is not sufficient to activate VWF,5 the primary autoinhibitory element should be close to the A1 domain. A number of studies have demonstrated the importance of the O-glycosylated sequences flanking the A1 domain to its platelet-binding activity.6-12 Once A1 and LBD bind together, these flanking sequences may also modulate the unusual mechanical response of the A1-LBD interaction under certain shear flow conditions,13-15 adding another layer of complexity to the regulation of VWF activity.
Recent biophysical studies indicate that an autoinhibitory module (AIM) consisting of both N- and C-terminal A1-flanking sequences adopts a quasi-stable conformation that prevents A1 binding to LBD.16-18 Application of tension around 20 pN, which is achieved when the VWF multimer is placed under high shear,5 could disrupt the protection of the A1 domain by the AIM to enable A1 binding to LBD.18 Such tension should not affect the conformational integrity of the A1 domain, because A1 is encompassed by a disulfide bond (Cys1272-Cys1458) that would require a rupture force of >100 pN.19 Consistent with the notion of a discontinuous AIM, removal of either N- or C-terminal part of the AIM from the recombinant AIM-A1 protein significantly increases its binding affinity for LBD.17,18 Moreover, introduction of type 2B von Willebrand disease mutations or desialylation of O-glycans in the AIM-A1 protein also destabilizes the AIM and increases the exposure of the GPIbα-binding site in the A1 domain.18,20,21 The inhibitory nanobody that comprises caplacizumab, referred to as VHH81, binds primarily to N-terminal AIM (NAIM) residues proximal to the A1 domain and significantly increases the rupture force for the AIM,18 suggesting that AIM may be a target for modulation of VWF activity.
Recent studies have disputed the notion of the discontinuous AIM.22,23 The controversy may stem partly from how the A1 domain is defined. In this study and our earlier reports, the A1 domain is defined as residues encompassed by the 1272-1458 disulfide bond. Sequences outside the disulfide bond but resolved in the crystal structures, such as residues 1264-1271 and 1459-1469,23 are not considered as a part of the A1 domain because they could be separated from A1 under tension, and they are not required for A1 binding to LBD.18 Instead, we consider them as a part of the AIM, defined as NAIM (residues 1238-1271) and C-terminal AIM (CAIM; residues 1459-1493). In comparison, other groups consider these residues as a part of the A1 domain, because they interact noncovalently with residues inside of the 1272-1458 disulfide bond and help stabilize the A1 domain. Residues outside of 1264-1469 were considered only as “linker” sequences, implying a lack of stable conformation or specific interactions therein.22,23 There are 8 O-glycans in the AIM or the “linker” sequences, at residues 1248, 1255, 1256, 1263, 1468, 1477, 1486, and 1487.24,25 Previous mutagenesis analyses of O-glycosylation sites around the A1 domain have identified T1255A and T1255A/T1256A as activating mutations.7,9 In addition, an activated VWF variant was generated when an unrelated O-glycosylated sequence was inserted between residues 1263 and 1264.12 The “linker” region, including glycans therein, was thought as unstructured, and it “simply shields the A1 domain and reduces its accessibility to platelet GPIbα.”22 Thus, at the center of the controversy is the role of residues 1238-1260 (ie, the distal portion of the NAIM) in modulating the platelet-binding activity of VWF and whether these “linker” residues contain any specific interactions with themselves or residues in the A1 domain.
In this study, we conducted targeted screens for anti-AIM nanobodies, and we have identified both activating and inhibitory nanobodies. The characterization of these nanobodies provides evidence for the presence of specific interactions within residues 1238-1260 and for the conformational integrity of the AIM that determines the VWF activity.
Methods
Materials and animals
Recombinant protein expression and purification
Biotinylated VWF proteins AIM-A1 (residues 1238-1493), NAIM-A1 (1238-1461), A1-CAIM (1268-1493), tAIM-A1 (1261-1472), related variants, and GPIbα-LBD have been described previously.18,28 VWF proteins Biotag-1238-1493-SpyTag, Biotag-1268-1493-SpyTag, and Biotag-1238-1461-SpyTag have been described previously.18 The DNA fragment encoding Biotag-1261-1472-SpyTag was similarly cloned and expressed.18 Constructs with a biotag (AviTag) were expressed from Expi293F BirA cells for biotinylation. Nonglycosylated AIM-A1 (Met-1238-1481-6His) was expressed in the cytoplasm of Shuffle T7 express cells (New England Biolabs). Nonglycosylated NAIM peptides with a N-terminal biotin group were synthesized by Genscript (supplemental Table 2, available on the Blood website). O-glycosylated NAIM peptides were expressed as Fc-fusion proteins in mammalian cells. Nanobodies were expressed in the cytoplasm of Escherichia coli and purified after cell lysis.18
Immunized yeast display library and targeted sorting
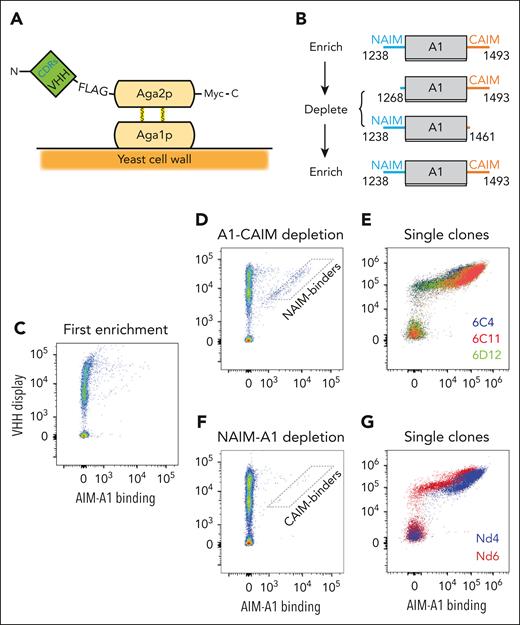
One adult Lama glama was immunized with the AIM-A1 protein by Abbiotec (Escondido, CA). Subsequently peripheral blood mononuclear cells were collected, from which complementary DNA of variable heavy domain of heavy chain (VHH) genes was amplified to construct a yeast display library. A library size of around 1.4 × 108 transformants was obtained from colony counts after serial dilutions of transformants. The library was initially enriched for AIM-A1 binding, then depleted with either NAIM-A1 or A1-CAIM, and again enriched for AIM-A1 binding (Figure 1). The details of this process are described in the supplemental Methods. Individual clones were then amplified and tested for their abilities to activate or inhibit the platelet-binding activity of VWF.
Screen of AIM-binding nanobodies by yeast surface display. (A) Schematic of the constructed yeast display library. Genes of VHH nanobodies were fused to that encoding the Aga2p subunit and VHH proteins were displayed on the yeast surface. (B) Schematic of the screening strategy. Yeasts were sorted for AIM-A1 binding, amplified, depleted with A1-CAIM or NAIM-A1 and re-sorted for AIM-A1 binding to obtain AIM-specific binders. Each protein is marked by the starting and ending VWF residues. (C-G) Flow cytometry plots showing binding of yeasts to AIM-A1 after various screening steps. In each plot, y-axis shows FLAG-tagged nanobody-Aga2p fusion protein expression on the yeast surface that is detected by Alexa Fluor 488-conjugated anti-FLAG antibody, and x-axis shows binding of the biotinylated AIM-A1 protein as detected by streptavidin-conjugated allophycocyanin. NAIM- and CAIM-specific binders were isolated from the approximate gates illustrated in the plots (D,F). After the second enrichment, individual clones (E,G) were isolated, amplified, and confirmed for AIM-A1 binding.
Screen of AIM-binding nanobodies by yeast surface display. (A) Schematic of the constructed yeast display library. Genes of VHH nanobodies were fused to that encoding the Aga2p subunit and VHH proteins were displayed on the yeast surface. (B) Schematic of the screening strategy. Yeasts were sorted for AIM-A1 binding, amplified, depleted with A1-CAIM or NAIM-A1 and re-sorted for AIM-A1 binding to obtain AIM-specific binders. Each protein is marked by the starting and ending VWF residues. (C-G) Flow cytometry plots showing binding of yeasts to AIM-A1 after various screening steps. In each plot, y-axis shows FLAG-tagged nanobody-Aga2p fusion protein expression on the yeast surface that is detected by Alexa Fluor 488-conjugated anti-FLAG antibody, and x-axis shows binding of the biotinylated AIM-A1 protein as detected by streptavidin-conjugated allophycocyanin. NAIM- and CAIM-specific binders were isolated from the approximate gates illustrated in the plots (D,F). After the second enrichment, individual clones (E,G) were isolated, amplified, and confirmed for AIM-A1 binding.
Platelet aggregometry
Written informed consent was obtained from participants before their inclusion in studies, and all procedures using donor-derived human blood and platelets were approved by the Institutional Review Board at Emory University. Platelet aggregation was performed largely as described.18
Biolayer interferometry
Biolayer interferometry experiments were performed on an Octet QKe instrument (ForteBio, Fremont, CA), largely as described previously.5
Hydrogen-deuterium exchange mass spectrometry (HDX-MS)
HDX-MS was performed largely as described.21
Parallel-plate flow chamber assay
Parallel-plate flow chamber experiments were performed using a Ibidi μ-Slide system. A tissue culture-treated Ibidi μ-Slide VI 0.1 device was coated with type 1 bovine collagen fibrils, and blocked with phosphate-buffered saline containing 1% bovine serum albumin. After citrated whole blood was mixed with 2 μg/mL DIOC-6 (Invitrogen), 2 mM CaCl2, and various antagonists for 30 minutes, it was perfused at desired shear rates to the chamber using Harvard Apparatus Pump 11 Elite. A total of 3 to 4 images were captured along the center of the chamber at desired time points in the fluorescein-5-isothiocyanate (FITC) channel. Each experiment was performed in 2 separate flow channels at randomly nonconsecutive shear rates. Area covered by adhered platelets was calculated as described before.18
X-ray crystallography
The AIM-A1/nanobody complexes were purified and concentrated to ∼16 mg/mL for crystallization. Crystals were harvested and 30% glycerol added as a cryo-protectant, then flash-frozen in liquid nitrogen for data collection on beamline i24 at Diamond Light Source. Diffraction data were collected from multiple crystals and processed to 3.05-Å resolution. The structure was solved using molecular replacement with the A1 domain crystal structure (1auq) and a nanobody model generated using AlphaFold.29,30 Manual model building was performed using COOT and refinement with PHENIX/Refmac5/pdb-redo (Table 1). Figures were generated using PyMOL.
Data collection and structure refinement statistics
| Sample . | AIM-A1 Nd6 complex . |
|---|---|
| Data collection | |
| Space group | P 1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 38.8, 174.3, 197.3 |
| α, β, γ (°) | 90, 89.96, 90 |
| Resolution (Å)∗ | 65.8-3.05 (3.34-3.05) |
| Rmerge† | 0.709 (2.778) |
| Rpim† | 0.0.142 (0.573) |
| Total number unique | 35 681 (1 758) |
| Mean(I)/sd(I) | 5.4 (1.5) |
| Completeness (spherical) (%) | 71.5 (15.0) |
| Completeness (ellipsoidal) (%) | 89.9 (39.4) |
| Multiplicity | 26.0 (23.1) |
| CC1/2‡ | 0.984 (0.375) |
| Wavelength | 0.9795 |
| Refinement | |
| No. reflections | 34 292 |
| Rwork§,∗ | 0.234 |
| Rfree | 0.297 |
| Average B factor (Å2) | |
| AIM-A1 chains A/B/E/G/ | 85/89/74/75 |
| Nd6 chains C/D/F/H/ | 50/50/47/43 |
| 4 × A2D A/B/E/G/ | 127/108/101/90 |
| 2 × CPS chains C/F | 66/84 |
| Solvent (75) | 42 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.604 |
| Ramachandran statistics‖ | |
| Favored (%) | 85.9 |
| Allowed (%) | 9.4 |
| Outliers (%) | 4.7 |
| Sample . | AIM-A1 Nd6 complex . |
|---|---|
| Data collection | |
| Space group | P 1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 38.8, 174.3, 197.3 |
| α, β, γ (°) | 90, 89.96, 90 |
| Resolution (Å)∗ | 65.8-3.05 (3.34-3.05) |
| Rmerge† | 0.709 (2.778) |
| Rpim† | 0.0.142 (0.573) |
| Total number unique | 35 681 (1 758) |
| Mean(I)/sd(I) | 5.4 (1.5) |
| Completeness (spherical) (%) | 71.5 (15.0) |
| Completeness (ellipsoidal) (%) | 89.9 (39.4) |
| Multiplicity | 26.0 (23.1) |
| CC1/2‡ | 0.984 (0.375) |
| Wavelength | 0.9795 |
| Refinement | |
| No. reflections | 34 292 |
| Rwork§,∗ | 0.234 |
| Rfree | 0.297 |
| Average B factor (Å2) | |
| AIM-A1 chains A/B/E/G/ | 85/89/74/75 |
| Nd6 chains C/D/F/H/ | 50/50/47/43 |
| 4 × A2D A/B/E/G/ | 127/108/101/90 |
| 2 × CPS chains C/F | 66/84 |
| Solvent (75) | 42 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.604 |
| Ramachandran statistics‖ | |
| Favored (%) | 85.9 |
| Allowed (%) | 9.4 |
| Outliers (%) | 4.7 |
R.m.s, root mean square; sd, standard deviation.
Values in parentheses are for highest-resolution shell.
Rmerge = Sum(h) [Sum(j) [I(hj) − <Ih>]/Sum(hj) <Ih> where I is the observed intensity and <Ih> is the average intensity of multiple observations from symmetry-related reflections calculated.
CC1/2 = Pearson correlation coefficient.
Rwork = Sum(h) ||Fo|h − |Fc|h|/Sum(h)|Fo|h, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree was computed as in Rwork, but only for (5%) randomly selected reflections, which were omitted in refinement, calculated using REFMAC.
Ramachandran statistics were calculated using Molprobity.
Additional details of the above methods, as well as those of enzyme-linked immunosorbent assay (ELISA), sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE), western blot, and dot blot can be found in the supplemental Methods.
Results
Nanobodies bind and activate human VWF
A llama was immunized with the AIM-A1 protein, and its VHH genes were amplified to construct a yeast display library (Figure 1). The initial 1.4 × 108 transformants of this library were enriched for binding to AIM-A1, depleted with binding to the NAIM-A1 or A1-CAIM protein, and then sorted again for binding to AIM-A1 (Figure 1B). From the screen with depletion by A1-CAIM, 3 anti-NAIM clones, designated 6C4, 6C11, and 6D12, were identified for their abilities to induce binding of AIM-A1 to recombinant LBD of GPIbα (Figure 1C-E). Only in the presence of AIM-A1 could yeast cells displaying these nanobodies bind LBD (supplemental Figure 1).
Nanobodies 6C4, 6C11, and 6D12 were subcloned for bacterial overexpression, and purified nanobodies were monomeric (supplemental Figure 2). When added to stirred human platelet-rich plasma, all 3 nanobodies dose-dependently induced platelet aggregation, the extent of which matched that induced by 1.5 mg/mL ristocetin (Figure 2A-D). The aggregation was inhibited by DNA aptamer ARC1172 that targets the A1 domain26 or antibody 11A8 that targets the LBD16 (supplemental Figure 3), confirming that nanobody-induced aggregation was dependent on VWF and GPIbα. The half maximal effective concentration (EC50) values for 6C4, 6C11, and 6D12 were ∼167, ∼100, and ∼57 nM, respectively (Figure 2D). Consistently, binding of plasma VWF to immobilized LBD could be detected by ELISA but only in the presence of a nanobody (Figure 2E; supplemental Figure 4). Overall, these nanobodies could bind specifically to human VWF and induce its binding of GPIbα and activation of platelets.
Activation of VWF by anti-NAIM nanobodies. (A-C) Platelet aggregation traces induced by addition of noted nanobodies. Human platelet-rich plasma was gently stirred, and, at 30 seconds, 6C4 (A), 6C11 (B), or 6D12 (C) was added to noted concentrations spanning from tens of nanomolar to 2 μM. Each concentration was tested in triplicate. Ristocetin was added in separate runs to 1.5 mg/mL, shown in black. Platelets stirred without addition of an agonist is shown in a gray trace. (D) Dose-aggregation curves were plotted for each concentration of each nanobody and fit to a 4-parameter dose-response curve. Percentage aggregation is the inverse of optical densities seen in panels A-C. Error bars are the standard deviation (SD) of the measurements. (E) ELISA binding isotherms of plasma VWF from normal pooled plasma (NPP) to immobilized GPIbα-LBD in the absence or presence of 500 nM noted nanobodies. Data are shown as mean ± SD (n = 3). Some error bars are not visible because they are smaller than the symbols.
Activation of VWF by anti-NAIM nanobodies. (A-C) Platelet aggregation traces induced by addition of noted nanobodies. Human platelet-rich plasma was gently stirred, and, at 30 seconds, 6C4 (A), 6C11 (B), or 6D12 (C) was added to noted concentrations spanning from tens of nanomolar to 2 μM. Each concentration was tested in triplicate. Ristocetin was added in separate runs to 1.5 mg/mL, shown in black. Platelets stirred without addition of an agonist is shown in a gray trace. (D) Dose-aggregation curves were plotted for each concentration of each nanobody and fit to a 4-parameter dose-response curve. Percentage aggregation is the inverse of optical densities seen in panels A-C. Error bars are the standard deviation (SD) of the measurements. (E) ELISA binding isotherms of plasma VWF from normal pooled plasma (NPP) to immobilized GPIbα-LBD in the absence or presence of 500 nM noted nanobodies. Data are shown as mean ± SD (n = 3). Some error bars are not visible because they are smaller than the symbols.
Activating nanobodies bind to a glycosylated distal sequence in NAIM
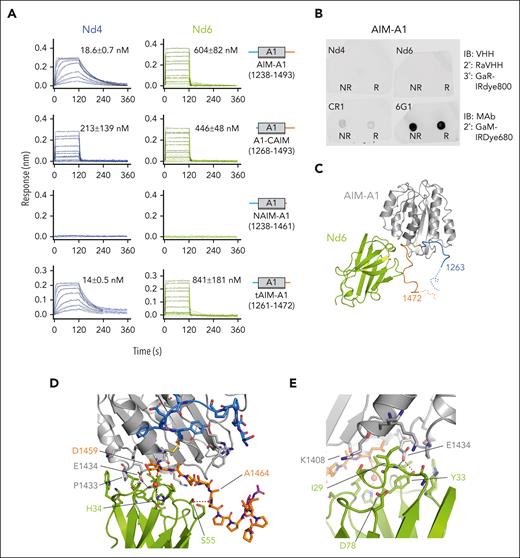
To define the mechanism by which these nanobodies activate human VWF, we characterized their binding epitopes. 6C4 bound with the lowest affinity (120 nM) among the 3 clones to AIM-A1. In comparison, 6C11 and 6D12 bound with tighter affinities at 14 and 31 nM, respectively (Figure 3A; supplemental Table 3). All 3 nanobodies exhibited binding to the NAIM-A1 protein but no detectable binding to A1-CAIM and tAIM-A1 (residues 1261-1472), confirming that these nanobodies target residues in the NAIM (Figure 3A;supplemental Figure 4C), likely residues preceding 1261.
Identification of the epitopes of VWF-activating nanobodies. (A) Representative BLI sensorgrams of immobilized AIM-A1, A1-CAIM, NAIM-A1, or tAIM-A1 binding to a serial dilution of purified nanobody 6C4 (blue), 6C11 (red), or 6D12 (green). Nanobody concentration ranges are 2 μM to 2 nM for 6C4 binding to AIM-A1, 250 nM to 2 nM for 6C11 and 6D12 binding to AIM-A1, and 500 nM to 1 nM for all others. The fitted affinity, when obtained, is shown in each panel. For AIM-A1, association was performed for 180 seconds and dissociation for 360 seconds. For A1-CAIM, NAIM-A1, and tAIM-A1, association was performed for 80 seconds and dissociation for 200 seconds. The fitting trace for each concentration is shown in black. (B) Western blots of nonreduced (NR) or reduced (R) AIM-A1 with noted nanobody or antibody 6G1. To accommodate the fast off-rate of 6C4, a biotinylated rabbit anti-VHH antibody (bio-RaVHH) and streptavidin-IRDye680 were used to detect blotting of AIM-A1 by 6C4. After blotting with 6C11 and 6D12, a rabbit anti-VHH antibody (RaVHH) and fluorescent goat anti-rabbit antibody (GaR) were used to visualize the blots. 6G1 binds to a linear epitope in the CAIM, and it was detected using a fluorescent goat anti-mouse secondary antibody (GaM). (C) Dot blots of biotinylated NAIM peptides (10 ng per spot) and bio-AIM-A1 (1 μg per spot). Each membrane strip was blotted with noted nanobody separately and detected with RaVHH and fluorescent GaR. To verify proper adsorption of biotinylated peptides and bio-AIM-A1 protein to the strip, the same strip was blotted with streptavidin-IRDye680. (D) ELISA binding isotherms of 6D12 to immobilized glycosylated AIM-A1 purified from mammalian cells (orange circles) and nonglycosylated AIM-A1 purified from E coli (black squares). Data are shown as mean ± SD (n = 3). Responses were fit to a hyperbola and the KD (10.7 ± 3.2 nM) was derived from half-maximal binding. (E) BLI binding traces of 6D12, in a dilution series, to immobilized NAIM-Fc, 1253-1266-Fc, and neuraminidase-treated NAIM-Fc (asialo-NAIM-Fc). Global kinetic fitting at ratio 1:1 is shown in red, and kinetic KD is listed on top of the sensorgrams.
Identification of the epitopes of VWF-activating nanobodies. (A) Representative BLI sensorgrams of immobilized AIM-A1, A1-CAIM, NAIM-A1, or tAIM-A1 binding to a serial dilution of purified nanobody 6C4 (blue), 6C11 (red), or 6D12 (green). Nanobody concentration ranges are 2 μM to 2 nM for 6C4 binding to AIM-A1, 250 nM to 2 nM for 6C11 and 6D12 binding to AIM-A1, and 500 nM to 1 nM for all others. The fitted affinity, when obtained, is shown in each panel. For AIM-A1, association was performed for 180 seconds and dissociation for 360 seconds. For A1-CAIM, NAIM-A1, and tAIM-A1, association was performed for 80 seconds and dissociation for 200 seconds. The fitting trace for each concentration is shown in black. (B) Western blots of nonreduced (NR) or reduced (R) AIM-A1 with noted nanobody or antibody 6G1. To accommodate the fast off-rate of 6C4, a biotinylated rabbit anti-VHH antibody (bio-RaVHH) and streptavidin-IRDye680 were used to detect blotting of AIM-A1 by 6C4. After blotting with 6C11 and 6D12, a rabbit anti-VHH antibody (RaVHH) and fluorescent goat anti-rabbit antibody (GaR) were used to visualize the blots. 6G1 binds to a linear epitope in the CAIM, and it was detected using a fluorescent goat anti-mouse secondary antibody (GaM). (C) Dot blots of biotinylated NAIM peptides (10 ng per spot) and bio-AIM-A1 (1 μg per spot). Each membrane strip was blotted with noted nanobody separately and detected with RaVHH and fluorescent GaR. To verify proper adsorption of biotinylated peptides and bio-AIM-A1 protein to the strip, the same strip was blotted with streptavidin-IRDye680. (D) ELISA binding isotherms of 6D12 to immobilized glycosylated AIM-A1 purified from mammalian cells (orange circles) and nonglycosylated AIM-A1 purified from E coli (black squares). Data are shown as mean ± SD (n = 3). Responses were fit to a hyperbola and the KD (10.7 ± 3.2 nM) was derived from half-maximal binding. (E) BLI binding traces of 6D12, in a dilution series, to immobilized NAIM-Fc, 1253-1266-Fc, and neuraminidase-treated NAIM-Fc (asialo-NAIM-Fc). Global kinetic fitting at ratio 1:1 is shown in red, and kinetic KD is listed on top of the sensorgrams.
Similar to monoclonal antibody 6G1,27 all 3 nanobodies could recognize denatured AIM-A1 in western blot (Figure 3B), suggesting that they recognize a linear epitope in the NAIM. However, they could not recognize any 20-mer synthetic peptides covering the NAIM sequence on a dot blot, although they could recognize AIM-A1 on the same blot (Figure 3C), suggesting that their epitopes may contain an entity beyond the primary protein sequence.
The binding epitope of nanobody 6D12 was further defined. Recombinant AIM-A1 produced from Expi293F cells contains multiple O-linked core 1 glycans.20 To test the hypothesis that the 6D12 epitope may include an O-glycan, nonglycosylated AIM-A1 protein was produced from E coli (supplemental Figure 5A-B). As expected,27 antibody 6G1 exhibited the same binding to both glycosylated and nonglycosylated AIM-A1 (supplemental Figure 5C). In comparison, 6D12 bound only to glycosylated AIM-A1, with an apparent affinity of ∼10 nM (Figure 3D). Next, human IgG1-Fc fusion proteins that contained the NAIM sequence (residues 1238-1271), or residues 1253-1266 that contained 3 O-glycosylation sites, were expressed from Expi293F cells (supplemental Figure 6A). 6D12 exhibited tight binding to both NAIM-Fc and 1253-1266–Fc, with binding affinities of 3.2 and 4.3 nM, respectively (Figure 3E; supplemental Figure 6B). Overall, these results demonstrate that the 6D12 epitope is in residues 1253-1266 of the NAIM and contains part of O-glycans therein. Consistently, activation of plasma VWF by 6D12 was not affected by the polymorphism at residue 1472 in the CAIM (supplemental Figure 7).
Binding of 6D12 exposes the GPIbα-binding site in the A1 domain
To analyze how 6D12 would activate A1, conformational dynamics of AIM-A1 with and without 6D12 binding was measured and compared by HDX-MS (Figure 4; supplemental Figures 8 and 9). A total of 121 peptic fragments were identified in both 6D12-bound and apo states of AIM-A1, covering ∼97% of the sequence (supplemental Figure 8). A few fragments were identified covering VWF residues 1253-1266, but none exhibited substantial decrease in relative deuterium uptake (Figure 4B; supplemental Figure 8). This may be due to the inclusion of O-glycans in the 6D12 epitope. On the contrary, 6D12 binding increased HDX of several parts in the A1 domain, including particularly residues 1308-1313 (α1β2 loop) and 1328-1340 (β3α2 loop) that are a part of the GPIbα-binding site (Figure 4C-D). In addition, HDX of the nearby α3β4 loop (residues 1372-1378) was also significantly increased (Figure 4E). These results indicate that 6D12 binding increases the solvent accessibility of the GPIbα-binding site.
Comparative HDX of AIM-A1 protein with and without bound 6D12. (A) Relative deuterium uptake displayed on the 3D structure of the A1 domain and partial AIM (PDB 1SQ0). Residues experiencing more exchange in the presence of 6D12 are colored in shades of red, no difference in exchange is displayed as white, and decreased exchange is shown in cyan. Note that many residues in the AIM were unresolved in this structure. (B-E) Uptake plots of representative peptides in AIM-A1 with (red) and without (black) bound 6D12 at 10, 30, 100, and 1000 seconds of exchange. Peptide 1239-1252 (B) is a NAIM-peptide. Peptide 1308-1318 (C) is in the α1β2 loop of the A1 domain, peptide 1330-1340 (D) is in the β3α2 loop, and peptide 1369-1382 (E) is in the α3β4 loop. For each time point, experiments were performed in triplicate. ∗∗∗∗P < .0001 using a Student t test; ∗∗∗P < .001; ∗∗P < .01; ∗P < .05.
Comparative HDX of AIM-A1 protein with and without bound 6D12. (A) Relative deuterium uptake displayed on the 3D structure of the A1 domain and partial AIM (PDB 1SQ0). Residues experiencing more exchange in the presence of 6D12 are colored in shades of red, no difference in exchange is displayed as white, and decreased exchange is shown in cyan. Note that many residues in the AIM were unresolved in this structure. (B-E) Uptake plots of representative peptides in AIM-A1 with (red) and without (black) bound 6D12 at 10, 30, 100, and 1000 seconds of exchange. Peptide 1239-1252 (B) is a NAIM-peptide. Peptide 1308-1318 (C) is in the α1β2 loop of the A1 domain, peptide 1330-1340 (D) is in the β3α2 loop, and peptide 1369-1382 (E) is in the α3β4 loop. For each time point, experiments were performed in triplicate. ∗∗∗∗P < .0001 using a Student t test; ∗∗∗P < .001; ∗∗P < .01; ∗P < .05.
Inhibition of VWF activity by anti-CAIM nanobodies
Two nanobodies, designated Nd4 and Nd6, were identified from the screen with depletion by NAIM-A1 to inhibit ristocetin-induced aggregation of human platelets (Figures 1F-G and 5A). In a parallel-plate flow chamber, both Nd4 and Nd6 significantly inhibited VWF-mediated platelet adhesion to collagen-coated surface under arterial shear rates (Figure 5B-C). Similar to that by nanobody VHH81, their inhibition was not complete, in that platelet adhesion was still visible at high shear rates. In comparison, ARC1172, a DNA aptamer that directly inhibits the VWF-GPIbα interaction, achieved complete inhibition regardless of the shear rates. We showed earlier that VHH81 inhibits VWF binding to GPIbα indirectly by binding primarily to the proximal portion of the NAIM and stabilizing the AIM.18 These results suggest that Nd4 and Nd6 likely inhibit VWF binding to GPIbα via a similar mechanism as VHH81.
Nanobodies Nd4 and Nd6 inhibit the activity of human VWF. (A) Ristocetin-induced platelet aggregation traces with or without the addition of 1 μM Nd4 or Nd6. Human platelet-rich plasma (PRP) was gently stirred with addition of each nanobody at 30 seconds, followed by addition of 1.5 mg/mL ristocetin at 60 seconds. (B) Representative flow chamber surface images at 120 seconds of human whole blood perfusion on collagen. Recalcified whole blood labeled with DIOC-6 was mixed with noted nanobody or aptamer inhibitors and then perfused over a collagen surface at given shear rates. The scale bar is 100 μm. Platelet adhesion was measured by thresholding pixel densities to calculate the area covered by adherent platelets. (C) Quantitative plots of platelet adhesion on collagen under noted shear rates and with treatment of various inhibitors (Nd4 in blue, Nd6 in green, VHH81 in pink, and ARC1172 in purple). Comparison between platelet adhesion was analyzed with a 2-way ANOVA with mixed effects with Tukey multiple comparison correction. Asterisks in given colors represent statistically significant values (P < .0001) between control blood and individual treatments at a given shear rate after multiple comparison corrections. Data are means ± SD (n = 8-12 fields analyzed per condition). Interaction F (8, 142) = 26.31; P < .0001; shear rate F (2, 142) = 7.47; P = .0008; treatment F (4, 142) = 141.7; P < .0001. ANOVA, analysis of variance.
Nanobodies Nd4 and Nd6 inhibit the activity of human VWF. (A) Ristocetin-induced platelet aggregation traces with or without the addition of 1 μM Nd4 or Nd6. Human platelet-rich plasma (PRP) was gently stirred with addition of each nanobody at 30 seconds, followed by addition of 1.5 mg/mL ristocetin at 60 seconds. (B) Representative flow chamber surface images at 120 seconds of human whole blood perfusion on collagen. Recalcified whole blood labeled with DIOC-6 was mixed with noted nanobody or aptamer inhibitors and then perfused over a collagen surface at given shear rates. The scale bar is 100 μm. Platelet adhesion was measured by thresholding pixel densities to calculate the area covered by adherent platelets. (C) Quantitative plots of platelet adhesion on collagen under noted shear rates and with treatment of various inhibitors (Nd4 in blue, Nd6 in green, VHH81 in pink, and ARC1172 in purple). Comparison between platelet adhesion was analyzed with a 2-way ANOVA with mixed effects with Tukey multiple comparison correction. Asterisks in given colors represent statistically significant values (P < .0001) between control blood and individual treatments at a given shear rate after multiple comparison corrections. Data are means ± SD (n = 8-12 fields analyzed per condition). Interaction F (8, 142) = 26.31; P < .0001; shear rate F (2, 142) = 7.47; P = .0008; treatment F (4, 142) = 141.7; P < .0001. ANOVA, analysis of variance.
Both Nd4 and Nd6 could bind to AIM-A1, albeit with disparate affinities of 19 and 600 nM, respectively (Figure 6A; supplemental Figure 10). Their binding specificity to CAIM was confirmed because neither nanobody exhibited binding to NAIM-A1. Both could bind to tAIM-A1 (residues 1261-1472) with similar affinities to AIM-A1, suggesting that their epitopes are proximal to A1. Unlike activating nanobodies described above, neither Nd4 nor Nd6 could detect AIM-A1 in dot blots, indicating that they target conformationally specific epitopes (Figure 6B). For comparison, antibody 6G1 that binds to residues 1461-1472 in the CAIM could blot AIM-A1, and CR1 that has a conformationally sensitive epitope could blot AIM-A1 weakly.27
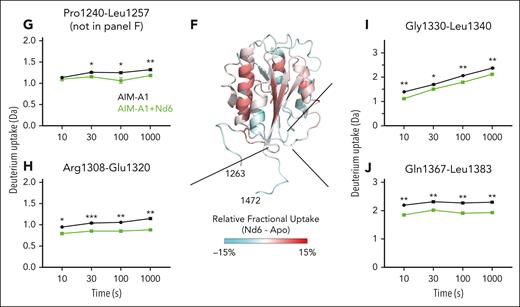
Epitopes of inhibitory nanobodies Nd4 and Nd6. (A) Representative BLI sensorgrams of immobilized AIM-A1, A1-CAIM, NAIM-A1, or tAIM-A1 binding to a serial dilution of purified nanobody Nd4 (blue) or Nd6 (green). Nanobody concentration ranges are 64 to 1 nM for Nd4 binding to AIM-A1, tAIM-A1, 4 μM to 2 nM for Nd4 binding to A1-CAIM, 20 μM to 20 nM for Nd6 binding to AIM-A1, A1-CAIM and tAIM-A1, and 2 μM to 125 nM for Nd4 and Nd6 binding to NAIM-A1. The fitted affinity, when obtained, is shown in each panel. For binding, association was performed for 120 seconds, and dissociation for 240 seconds. The fitting trace for each concentration is shown in black. (B) Dot blots of immobilized NR and R AIM-A1 by Nd4 and Nd6. 6G1, which recognizes a linear epitope, and CR1, which is partially conformationally sensitive, were included for comparison. (C) Crystal structure of the AIM-A1/Nd6 complex, shown in ribbon diagram. The nanobody Nd6 is shown in green, the A1 domain in gray, NAIM residues in sky blue, and CAIM residues in orange. Dashed lines denote unresolved residues in AIM-A1. (D-E) Close-up diagrams showing specific contacts between AIM-A1 and Nd6. Hydrogen bonds between side chains or main chains of noted residues are marked by dashed lines. Note that CAIM residues beyond residue Ala1464 do not show any direct contacts with Nd6. The GalNAC group on residue 1468 is colored magenta. (F) Relative deuterium uptake displayed on the 3D structure of the A1 domain and partial AIM as in the Nd6 complex. Residues experiencing more exchange in the presence of Nd6 are colored in shades of red, no difference in exchange is displayed as white, and decreased exchange is shown in cyan. Note that many residues in the AIM were unresolved in this structure. (G-J) Uptake plots of representative peptides in AIM-A1 with (green) and without (black) bound 6D12 at 10, 30, 100, and 1000 seconds of exchange. Peptide 1240-1257 (G) is a NAIM-peptide. Peptide 1308-1320 (H) is in the α1β2 loop of the A1 domain, peptide 1330-1340 (I) is in the β3α2 loop, and peptide 1367-1383 (J) is in the α3β4 loop. For each time point, experiments were performed in triplicate. ∗∗∗P < .001 using a Student t test; ∗∗P < .01; ∗P < .05.
Epitopes of inhibitory nanobodies Nd4 and Nd6. (A) Representative BLI sensorgrams of immobilized AIM-A1, A1-CAIM, NAIM-A1, or tAIM-A1 binding to a serial dilution of purified nanobody Nd4 (blue) or Nd6 (green). Nanobody concentration ranges are 64 to 1 nM for Nd4 binding to AIM-A1, tAIM-A1, 4 μM to 2 nM for Nd4 binding to A1-CAIM, 20 μM to 20 nM for Nd6 binding to AIM-A1, A1-CAIM and tAIM-A1, and 2 μM to 125 nM for Nd4 and Nd6 binding to NAIM-A1. The fitted affinity, when obtained, is shown in each panel. For binding, association was performed for 120 seconds, and dissociation for 240 seconds. The fitting trace for each concentration is shown in black. (B) Dot blots of immobilized NR and R AIM-A1 by Nd4 and Nd6. 6G1, which recognizes a linear epitope, and CR1, which is partially conformationally sensitive, were included for comparison. (C) Crystal structure of the AIM-A1/Nd6 complex, shown in ribbon diagram. The nanobody Nd6 is shown in green, the A1 domain in gray, NAIM residues in sky blue, and CAIM residues in orange. Dashed lines denote unresolved residues in AIM-A1. (D-E) Close-up diagrams showing specific contacts between AIM-A1 and Nd6. Hydrogen bonds between side chains or main chains of noted residues are marked by dashed lines. Note that CAIM residues beyond residue Ala1464 do not show any direct contacts with Nd6. The GalNAC group on residue 1468 is colored magenta. (F) Relative deuterium uptake displayed on the 3D structure of the A1 domain and partial AIM as in the Nd6 complex. Residues experiencing more exchange in the presence of Nd6 are colored in shades of red, no difference in exchange is displayed as white, and decreased exchange is shown in cyan. Note that many residues in the AIM were unresolved in this structure. (G-J) Uptake plots of representative peptides in AIM-A1 with (green) and without (black) bound 6D12 at 10, 30, 100, and 1000 seconds of exchange. Peptide 1240-1257 (G) is a NAIM-peptide. Peptide 1308-1320 (H) is in the α1β2 loop of the A1 domain, peptide 1330-1340 (I) is in the β3α2 loop, and peptide 1367-1383 (J) is in the α3β4 loop. For each time point, experiments were performed in triplicate. ∗∗∗P < .001 using a Student t test; ∗∗P < .01; ∗P < .05.
Structural and dynamic characterization of the AIM-A1/Nd6 complex
The crystal structure of AIM-A1 in complex with Nd6 was determined to 3.0-Å resolution (supplemental Figures 11 and 12; Figure 6C-E; Table 1). The AIM-A1/Nd4 complex was also prepared for crystallization (supplemental Figure 12), but the crystal contained only Nd4, likely due to the acidic crystallizing condition that may have disrupted the complex (data not shown). In the Nd6 complex structure, VWF residues 1263 to 1472 were resolved, including the O-linked N-acetylgalactosamine on residue 1468 (Figure 6C-D). Nd6 interacts with the proximal portion of the CAIM and some residues from the A1 domain. Specifically, the Nd6 Ile29 main chain nitrogen and Tyr33 side chain hydroxyl form hydrogen bonds with Glu1434 of VWF, and the Nd6 His34 side chain forms a water-mediated hydrogen bond with the main chain of Asp1459 of VWF (Figure 6D-E). In addition, there is a hydrogen bond interaction formed by Nd6 Ser55 side chain hydroxyl and the Ala1464 main chain carbonyl of VWF, and a long-range salt bridge formed by Asp78 of Nd6 and Lys1408 of VWF. The rest of interactions between VWF and Nd6 are van der Waals contacts, consistent with the relatively weak binding affinity between Nd6 and AIM-A1.
HDX of AIM-A1 in complex with equal molar Nd6 was measured and compared with apo-AIM-A1. A total of 104 peptic fragments were identified in both states of AIM-A1 (supplemental Figures 13 and 14). Binding of Nd6 reduced HDX of several parts in the AIM-A1 protein, including residues in the NAIM, α1β2, β3α2, and α3β4 loops (Figure 6F-J). This is opposite of the effects induced by 6D12 binding (Figure 4). Binding of Nd4 and VHH81 exhibited similar reducing effects on the same regions (supplemental Figures 15 and 16), suggesting that these inhibitory nanobodies stabilize the AIM and reduce the exposure of the GPIbα-binding site.
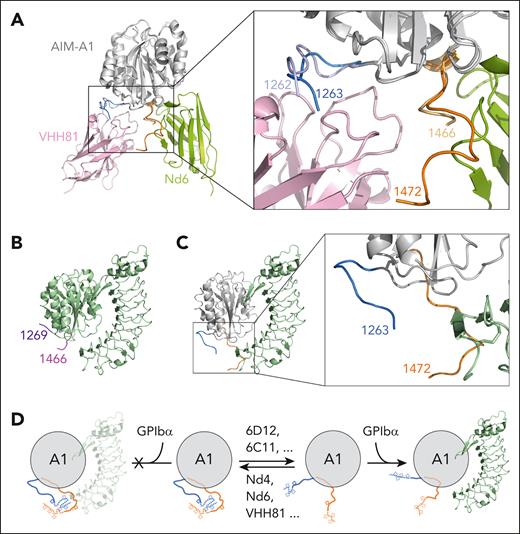
A comparison of Nd6 and VHH81 complex structures18 reveals that the A1 domain structure is indistinguishable (Figure 7A). Although Nd6 and VHH81 bind to drastically different epitopes in AIM-A1, AIM residues in these 2 structures take on similar conformations, which suggests that both nanobodies stabilize the same conformation of the AIM. In contrast, 2 major differences are apparent between AIM-A1 structures in their complexes with Nd6 and LBD.31-33 One lies in the α1β2 loop (supplemental Figure 17). The other, larger difference lies in the AIM, both in terms of resolved residues and their conformations (Figure 7B-C). When superimposed together, CAIM residues and the α1β2 loop in the Nd6 complex would overlap in space with the N-terminal residues in the LBD, illustrating the structural basis for Nd6 inhibition.
Comparison of the Nd6 complex structure with other A1 structures suggests a model of VWF autoinhibition and activation. (A) Overlaid Nd6 and VHH81 complex structures, with resolved AIM residues zoomed in and highlighted in the box on the right. Nd6 is shown in green, VHH81 in pink, and both AIM-A1 in gray. Resolved NAIM and CAIM residues from the Nd6 structure are shown in sky blue and orange, respectively. Resolved NAIM and CAIM residues from the VHH81 structure are shown in light purple and yellow, respectively. (B) Ribbon diagram of the A1/LBD complex structure (PDB: 1sq0), with the resolved AIM residues highlighted in purple and magenta. (C) Overlaid Nd6 and LBD complex structures, showing only AIM-A1 from the Nd6 complex structure and LBD from the LBD complex structure. The highlight box on the right shows the overlap of resolved AIM residues with the N-terminus of the LBD. (D) Illustration of the AIM model of VWF autoinhibition and activation. A structured AIM would have steric hindrance with the LBD, thus shielding A1 from binding to LBD. Conformationally insensitive nanobodies, such as 6D12 and 6C11, can bind to the NAIM residues distal from the A1 domain and disrupt the AIM, which exposes A1 for binding to the LBD. Conversely, conformationally sensitive nanobodies such as Nd6 and VHH81 can bind and stabilize the AIM, thus inhibiting A1 binding to the LBD.
Comparison of the Nd6 complex structure with other A1 structures suggests a model of VWF autoinhibition and activation. (A) Overlaid Nd6 and VHH81 complex structures, with resolved AIM residues zoomed in and highlighted in the box on the right. Nd6 is shown in green, VHH81 in pink, and both AIM-A1 in gray. Resolved NAIM and CAIM residues from the Nd6 structure are shown in sky blue and orange, respectively. Resolved NAIM and CAIM residues from the VHH81 structure are shown in light purple and yellow, respectively. (B) Ribbon diagram of the A1/LBD complex structure (PDB: 1sq0), with the resolved AIM residues highlighted in purple and magenta. (C) Overlaid Nd6 and LBD complex structures, showing only AIM-A1 from the Nd6 complex structure and LBD from the LBD complex structure. The highlight box on the right shows the overlap of resolved AIM residues with the N-terminus of the LBD. (D) Illustration of the AIM model of VWF autoinhibition and activation. A structured AIM would have steric hindrance with the LBD, thus shielding A1 from binding to LBD. Conformationally insensitive nanobodies, such as 6D12 and 6C11, can bind to the NAIM residues distal from the A1 domain and disrupt the AIM, which exposes A1 for binding to the LBD. Conversely, conformationally sensitive nanobodies such as Nd6 and VHH81 can bind and stabilize the AIM, thus inhibiting A1 binding to the LBD.
Discussion
In this study, a screen of nanobodies that specifically target the AIM instead of the A1 domain produced both activators and inhibitors of VWF (Figures 1, 2, and 5). All activating nanobodies are conformationally insensitive, and their epitopes were mapped to an O-glycosylated sequence in the NAIM that is distal to the A1 domain (Figure 3). Binding of the representative clone 6D12 induces the exposure of the GPIbα-binding site in the A1 domain (Figure 4). On the contrary, both inhibitory nanobodies are conformationally sensitive, and their epitopes include residues in the CAIM (Figure 6). Their binding reduces the exposure of the GPIbα-binding site (Figure 6). The AIM-A1/Nd6 complex structure resolves the most CAIM residues among all reported A1 structures, and these residues would overlap with bound GPIbα, suggesting that Nd6 binding stabilizes the AIM conformation to prevent A1 binding to GPIbα. Taken together, our results support a model for VWF autoinhibition and activation in which conformational stability of the AIM determines the platelet-binding activity of VWF (Figure 7D). In this model, NAIM and CAIM residues together form the AIM. The AIM masks the GPIbα-binding site but not the β3 strand in the A1 domain, which could be strengthened by VWF inhibitors or disrupted by activators. Similar to Nd6, VHH81 is also conformationally sensitive and inhibits VWF, although it binds primarily NAIM residues (Figure 7A).18 Similar to 6D12, 6G1 is a conformationally insensitive antibody that could activate the AIM-A1 protein.18 Unlike 6D12, 6G1 binds a linear epitope in the proximal portion of the CAIM.27
A key observation of the AIM-A1/Nd6 complex structure is that, although no direct contacts were observed between Nd6 and VWF residues 1465-1472, residues 1467-1472 were resolved for the first time in a crystal structure. Resolved residues of NAIM and CAIM point toward each other and away from the A1 domain (Figure 7A-C), likely placing some unresolved residues in the structure in the same vicinity for interaction with each other. The adoption of a specific conformation or interaction by the unresolved residues, especially 1253 to 1263 including the associated glycans, is supported by our discovery of activating nanobodies. If residues 1238-1260 did not contain a structural element and were only to provide hindrance as a “linker” sequence, attaching a sizable nanobody such as 6D12 to this stretch of residues would only increase its hindrance on GPIbα binding. However 6D12 binding clearly activates VWF and exposes its GPIbα-binding site. Our result is consistent with earlier reports that mutations at residues 1255 and/or 1256 could activate VWF.7,9
That NAIM and CAIM residues point toward each other has not been observed in previously reported structures of the A1 domain.26,29,31-41 Except for complex structures of A1 with LBD and AIM-A1 with VHH81,18,31 all previous A1 structures used nonglycosylated proteins produced from bacteria.26,29,32-41 In the first A1/LBD complex structure, a yeast-expressed protein containing residues 1261-1468 and bearing the R1306Q mutation was used, thus without the distal portion of the NAIM.31 The AIM-A1/VHH81 structure used the same glycosylated AIM-A1 protein as in the Nd6 complex structure,18 but not as many CAIM residues were resolved, presumably without stabilization of the CAIM by Nd6. Desialylation, particularly that of O-glycans around the A1 domain, contributes to VWF activation.20,42 Thus, a bacterial-expressed nonglycosylated A1 protein may not contain a sufficiently stable AIM. It may also support a nonnative conformation in the crystal structure. For instance, in the unbound A1 structure,29 the side chain of Ser1263 was pointing toward the A1 domain. If the residue was O-glycosylated as in its native form, its pointing toward A1 would have created steric hindrance. Several cryoEM structures of D1-A1 domains have been reported to model the packed VWF tubule in the Weibel-Palade body.43,44 Although proteins in these structures were produced from mammalian cells, they contain no CAIM residues beyond residue 1464. Neither structures show resolved NAIM residues before 1263.43,44 The E3 module in the D’D3 assembly that adjoins the NAIM is not resolved in these structures.
Despite stabilization by Nd6, the distal portion of the AIM, including O-glycans therein, is not resolved in the Nd6 complex structure. This is possibly due to the heterogeneity of O-glycans in the AIM. Although the AIM-A1 protein contains primarily core 1 O-glycans, both monosialylated and disialylated forms with either α-2,3 or α-2,6 linkage are present.20 At the present time, there are no effective means to generate the AIM-A1 protein with homogeneous O-glycans. The specific interactions involving sialylated residues 1253-1266 remain to be determined.
It was suggested that hydrogen bonds between proximal NAIM residues (eg, 1264-1271) and the A1 domain are critical in keeping the latter in an inactive conformation and breaking them induces A1 to an active conformation primed for LBD binding.23,45 Although proximal NAIM residues in Nd6 and VHH81 complex structures take on similar conformations (Figure 7A), they are sufficiently different to form different hydrogen bonds with A1. It is not apparent how different sets of hydrogen bonds between NAIM and A1 help upkeep the same autoinhibited state of A1.
Ristocetin is widely used as an activator of human VWF in research and diagnosis. The ristocetin-binding site in VWF include primarily the A1-proximal portion of the CAIM.16 Ristocetin forms dimers when exceeding a concentration of 1.1 mg/mL, and dimeric ristocetin may facilitate VWF-dependent agglutination of platelets.46 Ristocetin could also bind and flocculate many other plasma proteins such as fibrinogen.46 Nanobody 6D12 is much more specific for VWF with a nanomolar affinity than ristocetin (Figure 3). Because 6D12 remains monomeric for all the concentrations used in this study (supplemental Figure 2), the ability of dimerization or oligomerization may not be required for an activator of VWF.
6D12 exhibits a higher affinity for NAIM-Fc and 1253-1266–Fc than it does for AIM-A1 protein (Figures 2 and 4), suggesting that its epitope in the NAIM is somewhat protected in AIM-A1. Relatedly, although 6C11 exhibits a higher binding affinity for AIM-A1 than 6D12, the latter is superior in inducing platelet aggregation with a lower EC50 value. Thus, additional factors in full-length VWF may affect the accessibility of the NAIM, and each nanobody may activate VWF to a different extent. Future studies are needed to elucidate the intrinsic efficacy of these nanobodies as VWF activators.
Several nanobodies have been developed to target VWF for various purposes. Nanobody AU/VWFa-11 binds preferentially to activated A1 domain of VWF than its inactive state, and it does not affect the VWF-GPIbα interaction.47 Another nanobody with similar preferential binding to VWF containing type 2B von Willebrand disease mutations than wild-type VWF was reported recently.48 The epitopes of these nanobodies were not reported, and it is unclear how they specifically recognize the GPIbα-binding state of the A1 domain. In addition, Aymé et al reported an inhibitory nanobody targeting the A1 domain, and it cross-reacted with both human and mouse VWF.49 How it inhibits the A1 activity, by binding directly to the GPIbα-binding site or binding to the AIM like VHH81 and Nd6 do, is not clear. Kizlik-Masson et al developed a nanobody that binds the A3 domain, and its binding could be reduced by ADAMTS13 cleavage of the A2 domain nearby.50 Recently, de Maat et al reported 2 nanobodies targeting the D’D3 assembly and the CT/CK domain, respectively, both of which were used in a fusion protein to facilitate protease cleavage of VWF multimers as potential therapeutics.51 In this paper, we report several nanobodies, particularly 6D12, with an activating function. Compared with ristocetin, these nanobodies are monomeric, highly specific to VWF, and their activation of VWF is not affected by common polymorphisms in VWF. They may be useful in future investigation and measurement of physiological and pathophysiological processes involving activated VWF.
Acknowledgments
The authors thank Lisa Bixby and Aaron Rae of the Emory Pediatric/Winship Flow Cytometry Core for their assistance with flow cytometry sorting of yeasts and Michael Berndt for sharing CR1 and 6G1 antibodies. The authors acknowledge Diamond Light Source for time on Beamline I24 under Proposal 19880.
This work was supported in part by the National Institutes of Health (NIH), National Heart, Lung, and Blood Institute (NHLBI) grants HL082808, HL143794, and HL166654, and a grant from Hemophilia of Georgia that supports the VWD program at Children’s Healthcare of Atlanta; supported in part by NIH/NHLBI fellowship grants HL154656 (N.A.A.) and HL149357 (E.R.L.); in part by China Scholarship Council 202106235016, Guangci Visiting Scholar Program (2019; Q.L.) and a Judith Graham Pool Postdoctoral Research Fellowship from the National Hemophilia Foundation (Q.L.).
Authorship
Contribution: N.A.A., J.E., and R.L. designed the study; N.A.A., Z.M.-L., Q.L., E.R.L., A.J.S., M.S.W., and P.L. performed experiments; G.D. and R.F.S. collected clinical samples; N.A.A., Z.M.-L., Q.L., S.N., E.R.L., P.L., J.E., and R.L. analyzed results and prepared figures; N.A.A. and R.L. wrote the manuscript; and Q.L., S.N., E.R.L., J.E., and P.L. edited the manuscript.
Conflict-of-interest disclosure: R.L., N.A.A., and Q.L. are inventors on pending patents describing VWF-modulating nanobodies and their uses. The remaining authors declare no competing financial interests.
Correspondence: Renhao Li, Department of Pediatrics, Emory University School of Medicine, 2015 Uppergate Drive NE, Room 440, Atlanta, GA 30322; email: renhao.li@emory.edu.
References
Author notes
All data associated with this study are available in the main text or the supplemental Materials. Crystallographic data and coordinates will be deposited in the Protein Data Bank.
Materials are freely available to academic and not-for-profit investigators upon request from the corresponding author, Renhao Li (renhao.li@emory.edu).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal