Abstract
PTPN11, which encodes the tyrosine phosphatase SHP2, is mutated in approximately 35% of patients with juvenile myelomonocytic leukemia (JMML) and at a lower incidence in other neoplasms. To model JMML pathogenesis, we generated knockin mice that conditionally express the leukemia-associated mutant Ptpn11D61Y. Expression of Ptpn11D61Y in all hematopoietic cells evokes a fatal myeloproliferative disorder (MPD), featuring leukocytosis, anemia, hepatosplenomegaly, and factor-independent colony formation by bone marrow (BM) and spleen cells. The Lin−Sca1+cKit+ (LSK) compartment is expanded and “right-shifted,” accompanied by increased stem cell factor (SCF)–evoked colony formation and Erk and Akt activation. However, repopulating activity is decreased in diseased mice, and mice that do engraft with Ptpn11D61Y stem cells fail to develop MPD. Ptpn11D61Y common myeloid progenitors (CMPs) and granulocyte-monocyte progenitors (GMPs) produce cytokine-independent colonies in a cell-autonomous manner and demonstrate elevated Erk and Stat5 activation in response to granulocyte-macrophage colony-stimulating factor (GM-CSF) stimulation. Ptpn11D61Y megakaryocyte-erythrocyte progenitors (MEPs) yield increased numbers of erythrocyte burst-forming units (BFU-Es), but MEPs and erythrocyte-committed progenitors (EPs) produce fewer erythrocyte colony-forming units (CFU-Es), indicating defective erythroid differentiation. Our studies provide a mouse model for Ptpn11-evoked MPD and show that this disease results from cell-autonomous and distinct lineage-specific effects of mutant Ptpn11 on multiple stages of hematopoiesis.
Introduction
Juvenile myelomonocytic leukemia (JMML) is a rare, lethal myeloproliferative disorder (MPD) of early childhood, characterized by leukocytosis with prominent monocytosis, macrocytic anemia with fetal hemoglobinemia, hepatosplenomegaly, and selective hypersensitivity of hematopoietic progenitors to granulocyte-macrophage colony-stimulating factor (GM-CSF).1 Approximately 75% to 85% of JMML cases result from (typically mutually exclusive) gain-of-function mutations in NRAS, KRAS, or PTPN11 or homozygous loss-of-function mutations in NF1, which encodes a Ras GTPase-activating protein (RAS-GAP). All of these genes are components of the RAS/ERK signaling cascade, which, together with biochemical and pharmacologic analyses, implicates deregulation of this signaling pathway in JMML pathogenesis.2-5
Somatic mutations in PTPN11, which encodes the nonreceptor tyrosine phosphatase SHP2, are the most frequent cause of JMML, accounting for approximately 35% of cases. Somatic mutations are also found in a smaller percentage of acute leukemia, and rarely, in solid tumors.5 Germline mutations in PTPN11 cause approximately 40% to 50% of cases of Noonan syndrome (NS), a common autosomal dominant disorder characterized by facial abnormalities, proportionate short stature, and cardiac defects.6,7 NS patients frequently exhibit signs of MPD, most often in the form of self-limited leukocytosis/splenomegaly that resolves without sequelae and very rarely, JMML.1
SHP2 contains 2 SH2 domains (N-SH2, C-SH2), a catalytic (PTP) domain, and a C-terminal tail of unclear function. In its basal state, SHP2 activity is suppressed by intramolecular interactions between residues in the “backside loop” of the N-SH2 domain and the catalytic surface of the PTP domain.8,9 Most NS/leukemia PTPN11 mutations affect N-SH2 or PTP domain residues involved in basal inhibition, resulting in “activated mutants.” Somatic leukemia mutants typically show higher phosphatase activity compared with those associated with NS and NS/leukemia.5
Previous studies began to address the pathogenesis of SHP2-evoked MPD. Retroviral transduction of “activated” alleles of PTPN11 into murine bone marrow (BM) cells causes growth factor independence and GM-CSF and IL-3 hypersensitivity of myeloid progenitors.10-12 Such alleles also increase IL-3–evoked Erk, Akt, and Stat5 phosphorylation in mast cells12 and enhance GM-CSF–evoked Erk activation in bone marrow macrophages.10 Moreover, transplantation of BALB/c BM expressing the leukemia-associated alleles PTPN11E76K or PTPN11D61Y into lethally irradiated recipients causes a fatal MPD.12
Although these studies provided initial insights into the pathogenesis of SHP2-evoked MPD, several important questions remain. In the gene transduction/bone marrow transplantation (BMT) experiments, mutant PTPN11 was under retroviral promoter control, and therefore was not expressed at “appropriate” levels in all hematopoietic lineages. The resultant MPD is incompletely penetrant, mouse strain–specific, and often admixed with a T-cell lymphoma/leukemia (T-ALL) syndrome.11,12 Some mice receiving mutant PTPN11-transduced C57BL/6 BM cells develop anemia,11 a key feature of JMML, but recipients of mutant PTPN11-transduced BALB/c BM cells do not.12 Mice (on 129/Sv × C57BL/6 background) bearing a knockin of the NS/JMML allele Ptpn11D61G show sub-Mendelian inheritance due to incompletely penetrant cardiac defects, with only approximately 50% of expected Ptpn11D61G/+ mice born.13 Surviving Ptpn11D61G/+ mice develop MPD, but this disease is mild, indolent, and noninvasive.13 Conceivably, the effects of the Ptpn11D61G allele on hematopoiesis are obscured by genetic modifiers that permit survival, and could complicate data interpretation. Finally, the retroviral infection/BMT and knockin models preclude the examination of the cell-autonomous effects of leukemogenic Ptpn11 alleles expressed at physiologic gene dosage in different hematopoietic lineages.
To address these issues, we generated inducible knockin mice expressing the leukemogenic allele Ptpn11D61Y. Expression of Ptpn11D16Y evokes cell-autonomous and lineage-specific effects on multiple stages of hematopoiesis. Together, these result in a fatal and invasive MPD accompanied by anemia. Our model yields new insights into JMML pathogenesis and provides a tractable platform to investigate myeloid disorders initiated by oncogenic Ptpn11.
Methods
Mice
The D61Y mutation and a unique AgeI restriction site were introduced into the Ptpn11 locus by site-directed mutagenesis. The targeted mutated allele is rendered inactive by a STOP cassette flanked by loxP sites (Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Correctly targeted ES clones were identified and microinjected into C57BL/6 blastocysts. Mice derived from 2 clones that yielded high percentage chimeras were mated with C57BL/6 mice to generate F1 animals, which were crossed to Mx1-Cre transgenic mice (on C57BL/6 background) to generate Mx1-Cre;LSL-Ptpn11D61Y F2 mice. To induce the expression of Ptpn11D61Y, 3- to 4-week-old control and Mx1-Cre;LSL-Ptpn11D61Y mice were injected intraperitoneally with 300 μg polyinosinic-polycytidylic acid (pIpC; Amersham, Piscataway, NJ) every third day for 3 doses. Mice were monitored for disease and killed when moribund. LSL-Ptpn11D61Y mice were also crossed to ER-Cre mice (Tg(cre/Esr1)5Amc; The Jackson Laboratory, Bar Harbor, ME). All mouse studies were approved by the animal welfare committees of Harvard Medical School and University Health Network.
Flow cytometry, histology, and pathology examination
Flow cytometry was carried out as described in Document S1. Blood smears were stained with Wright-Giemsa. Complete blood counts were determined with a Hemavet 850FS (Drew Scientific, Dallas, TX). Tissues and organs were collected in 10% formalin and processed by the Specialized Histopathology Services at Brigham and Women's Hospital.
Colony assays
BM, spleen, or purified LSK or progenitor cells were suspended in methylcellulose medium (Stem Cell Technologies, Vancouver, BC) to assay for cytokine-independent colonies (M3234), granulocyte-macrophage colony-forming units (CFU-G/M/GM) (M3434), or erythroid burst-forming units (BFU-E) (M3234, 10 ng/mL IL-3 and 3 U/mL EPO), as indicated. Colonies were scored after 7 to 9 days. BFU-E colonies were stained with benzidine solution (0.4% benzidine [Sigma-Aldrich, St Louis, MO] in 12% acetic acid, and 0.3% H2O2). For some experiments, purified common myeloid progenitors (CMPs) and granulocyte-monocyte progenitors (GMPs) were cultured for 16 hours in media containing SCF (20 ng/mL) and IL-11 (10 ng/mL; R&D Systems, Minneapolis, MN) in the absence or presence of 4-hydroxytamoxifen (50 nM; Sigma-Aldrich), before replating in M3434 methylcellulose media. Purified megakaryocytic-erythroid progenitors (MEPs) or erythroid progenitors (EPs) were plated in M3334 media for 2 days for erythrocyte colony-forming unit (CFU-E) colonies or in M3434 for 8 days for BFU-E colonies.
Adoptive transfer
B6.SJL-Ptprca Pep3b/BoyJ CD45.1+ mice (The Jackson Laboratory) were irradiated with 2 doses of 4.75 gray each, then injected intravenously with BM or spleen cells from induced Ptpn11D61Y mice (C57BL/6, F7) together with 105 CD45.1 WT BM cells for radioprotection. Complete blood counts and the ratio of CD45.1+/CD45.2+ cells were monitored at the indicated times.
Statistical analysis
Data are presented as means plus or minus SEM, and were analyzed by 2-tailed Student t test. Some data were analyzed by Wilcoxon-Mann-Whitney test, as indicated.
Results
Ptpn11D61Y mice develop a fatal MPD
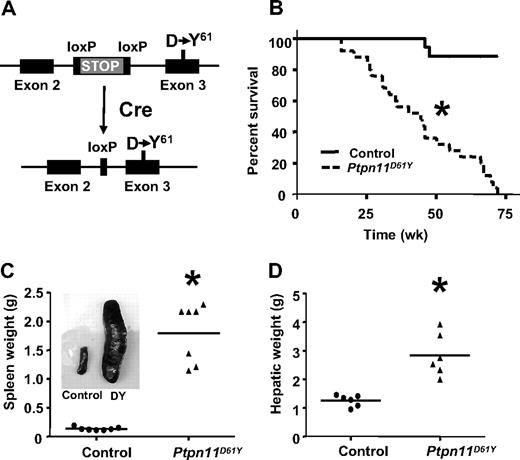
The 2 most common PTPN11 mutations in human leukemia are PTPN11E76K and PTPN11D61Y; their cognate proteins also have the highest degree of phosphatase activity among those SHP2 mutants tested in vitro.5,14 To better model human disorders caused by leukemogenic PTPN11 alleles, we introduced the D61Y mutation into the mouse Ptpn11 locus, along with a loxP-bracketed transcriptional “STOP” cassette (Figure 1A; Figure S1A-D). Properly targeted ES clones were injected into blastocysts, and germ-line transmission of the inducible Ptpn11D61Y allele (LSL-Ptpn11D61Y) was obtained (Figure S1E). When LSL-Ptpn11D61Y mice were crossed to EIIA-Cre transgenic mice, which express Cre at early stages of embryogenesis,15 there were no viable Ptpn11D61Y-expressing progeny (data not shown).
Mice expressing Ptpn11D61Y develop fatal MPD. (A) Schematic of LSL-Ptpn11D61Y allele with floxed “STOP” cassette (top) and expressing the Ptpn11D61Y mutation after Cre expression (bottom). (B) Kaplan-Meier analysis of a cohort of Ptpn11D61Y mice (n = 25) and littermate controls (n = 32). Cumulative survival was plotted against days after treatment with pIpC. Ptpn11D61Y mice have a median life span of 45 weeks. (C-D) Ptpn11D61Y mice develop splenomegaly (C) and hepatomegaly (D). (*P < .05.)
Mice expressing Ptpn11D61Y develop fatal MPD. (A) Schematic of LSL-Ptpn11D61Y allele with floxed “STOP” cassette (top) and expressing the Ptpn11D61Y mutation after Cre expression (bottom). (B) Kaplan-Meier analysis of a cohort of Ptpn11D61Y mice (n = 25) and littermate controls (n = 32). Cumulative survival was plotted against days after treatment with pIpC. Ptpn11D61Y mice have a median life span of 45 weeks. (C-D) Ptpn11D61Y mice develop splenomegaly (C) and hepatomegaly (D). (*P < .05.)
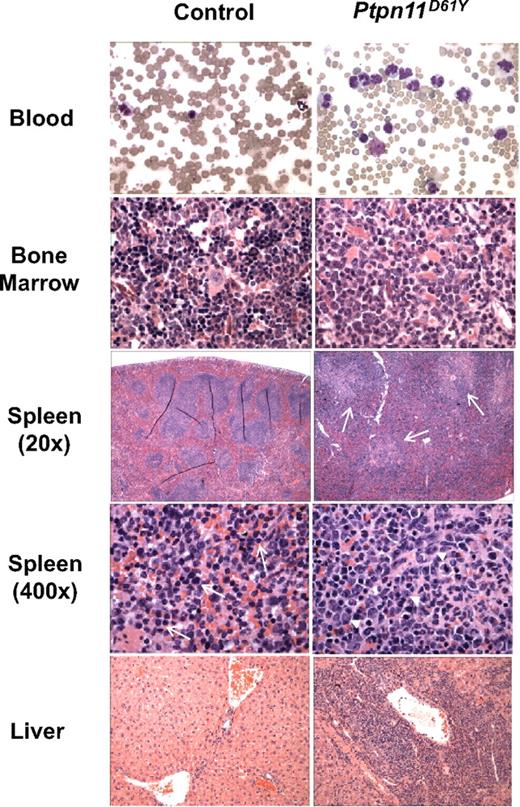
To circumvent the embryonic lethality of globally expressed Ptpn11D61Y and allow controlled, postnatal expression of this allele, LSL-Ptpn11D61Y mice were crossed to Mx1-Cre mice, which express Cre widely in response to double-strand (ds) RNA treatment.16 Three-week-old Mx1-Cre;LSL-Ptpn11D61Y/+ and control littermates (a mixture of Mx1-Cre;Ptpn11+/+, LSL-Ptpn11D61Y, or WT mice, all of which have no apparent phenotype; data not shown) were injected with pIpC (300 μg intraperitoneally) every third day for a total of 3 doses. Peripheral blood (PB), BM, and spleen cells, as well as hematopoietic stem cell (HSC)–enriched Lin−cKit+Sca1+(LSK) cells, common myeloid progenitors (CMPs), megakaryocytic-erythroid progenitors (MEPs), and granulocyte-macrophage progenitors (GMPs) from induced Mx1-Cre;LSL-Ptpn11D61Y (hereafter, Ptpn11D61Y) mice all showed complete deletion of the entire STOP cassette (Figure S1F,G). Although Ptpn11D61Y mice initially appeared healthy, all died prematurely, with a median survival of 45 weeks after injection (Figure 1B). Two control mice died of unknown causes; all others survived for more than 75 weeks without gross or histopathologic signs of disease (Figure 1B and data not shown). All Ptpn11D61Y mice developed MPD, characterized by progressive leukocytosis (granulocytosis and monocytosis) and hepatosplenomegaly (Figures 1C,D and 2; Table 1; and data not shown). Notably, in contrast to our retroviral/BMT model,12 Ptpn11D61Y animals developed significant anemia, although mutant red blood cells (RBCs) showed normal morphology and mean corpuscular volume (MCV; Figure 2, Table 1). Platelet counts in Ptpn11D61Y mice were in the normal range (Table 1). BM of diseased mice showed increased numbers of immature, predominantly granulocytic cells (Figure 2). Mutant spleens exhibited marked extramedullary hematopoiesis with disruption of the normal splenic architecture due to infiltration of mature myeloid cells into the red pulp (Figure 2). Flow cytometry revealed a 9- to 10-fold increase in the ratio of Mac1+Gr1+ cells and a 6- to 7-fold increase of Ter119+ erythroid progenitors in mutant spleens (Table 2). Although the percentage of B cells in mutant spleens was similar to that in control mice, there was a decrease in the relative number of splenic T cells (Table 2). Finally, periportal cuffing of liver sinusoids with infiltrating granulocytes was present in all diseased animals examined (Figure 2 and data not shown). Unlike knockin mice with induced expression of oncogenic KrasG12D,17 no lung adenomas, skin tumors, or other obvious neoplasms were detected in Ptpn11D61Y mice.
Histopathology of Ptpn11D61Y-evoked MPD.Ptpn11D61Y mice show marked increase of mature myeloid elements in the peripheral blood (×400), bone marrow (×400), and spleen (×20 and ×400), as well as myeloid infiltration in the liver (×100). Myeloid infiltration in the white pulp of Ptpn11D61Y spleen is indicated by arrows (spleen ×20). Whereas control spleens contain mainly immature RBCs (arrows, spleen ×400), mutant spleens show increased myeloid cells (arrowheads, spleen ×400).
Histopathology of Ptpn11D61Y-evoked MPD.Ptpn11D61Y mice show marked increase of mature myeloid elements in the peripheral blood (×400), bone marrow (×400), and spleen (×20 and ×400), as well as myeloid infiltration in the liver (×100). Myeloid infiltration in the white pulp of Ptpn11D61Y spleen is indicated by arrows (spleen ×20). Whereas control spleens contain mainly immature RBCs (arrows, spleen ×400), mutant spleens show increased myeloid cells (arrowheads, spleen ×400).
Ptpn11D61Y expression alters phenotypic HSC and progenitor compartments
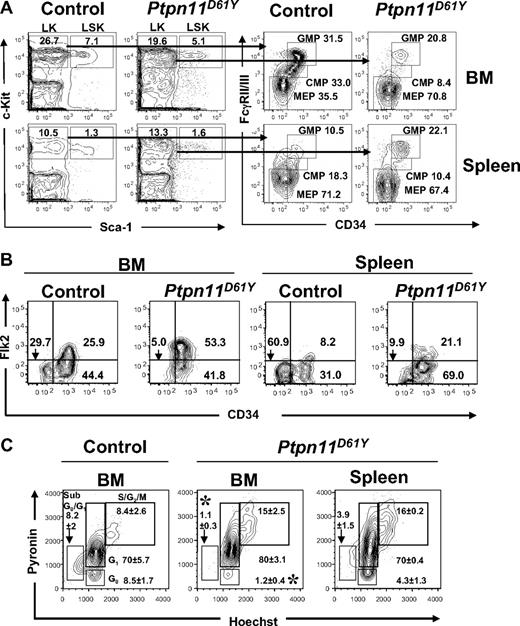
Next, we examined BM and splenic hematopoietic progenitor cells in Ptpn11D61Y mice in greater detail, using combinations of cell surface markers that, in normal BM, distinguish long-term (LT)–HSCs and short-term (ST)–HSCs from multipotent progenitors (MPPs), CMPs, common lymphoid progenitors (CLPs), MEPs, and GMPs. Mice with MPD showed decreased total BM cellularity and diminished BM Lin−Kit+Sca1− (LK) and LSK compartments (Figure 3A; Tables 2,3). Within the LSK compartment, the number of CD34−Flk2− cells (phenotypic LT-HSCs) was lower in mutant mice compared with control littermates. Mutant BM also had a lower proportion of CD150+CD48−LSK cells (n = 2; data not shown); in normal BM, this combination of markers identifies LT-HSCs with higher purity.18 Together, these data indicate a loss of phenotypic LT-HSCs in Ptpn11D61Y BM.
Abnormal distribution of phenotypic HSCs and myeloid precursors in the bone marrow and spleen of diseased Ptpn11D61Y mice. (A,B) Flow cytometric analysis of BM and splenic (A) LK subsets, including CMPs (LK-CD34+FcγRII/IIIlo), GMPs (LK-CD34+FcγRII/III+), and MEPs (LK-CD34−FcγRII/III−) and (B) LSK subsets, LT-HSCs (LSK-Flk2−CD34−), ST-HSCs (LSK-Flk2−CD34+), and MPPs (LSK-Flk2+CD34+). Representative contour plots are shown. All values are the mean frequency of the parental gate. See Tables 1 and 2 for absolute numbers of each population. (C) LSK cells from BM or spleen of control and Ptpn11D61Y mice were purified by FACS and stained with Hoechst 33342 (H), and Pyronin Y (PY). The mean percentages of cells in G0 (H−PY−), sub-G0G1 (H−PYlo), G1 (H−PY+), and S/G2M (H+PY+) are indicated. Representative contour plots are shown (n = 3; *P < .05).
Abnormal distribution of phenotypic HSCs and myeloid precursors in the bone marrow and spleen of diseased Ptpn11D61Y mice. (A,B) Flow cytometric analysis of BM and splenic (A) LK subsets, including CMPs (LK-CD34+FcγRII/IIIlo), GMPs (LK-CD34+FcγRII/III+), and MEPs (LK-CD34−FcγRII/III−) and (B) LSK subsets, LT-HSCs (LSK-Flk2−CD34−), ST-HSCs (LSK-Flk2−CD34+), and MPPs (LSK-Flk2+CD34+). Representative contour plots are shown. All values are the mean frequency of the parental gate. See Tables 1 and 2 for absolute numbers of each population. (C) LSK cells from BM or spleen of control and Ptpn11D61Y mice were purified by FACS and stained with Hoechst 33342 (H), and Pyronin Y (PY). The mean percentages of cells in G0 (H−PY−), sub-G0G1 (H−PYlo), G1 (H−PY+), and S/G2M (H+PY+) are indicated. Representative contour plots are shown (n = 3; *P < .05).
In contrast, the number of LSK cells was markedly increased in the mutant spleen. There was also a “rightward shift” within this compartment, with increased percentages of phenotypic ST-HSCs and MPPs (Table 3; Figure 3B). Overall, despite the smaller LSK compartment in the BM, Ptpn11D61Y mice produce an excess of total LSK cells and phenotypic LT-HSCs, most of which reside in the spleen.
Cell-cycle analysis using Hoechst 33342/Pyronin Y staining revealed an approximately 8-fold reduction in the percentage of G0 LSK cells in the BM of Ptpn11D61Y mice (Figure 3C). Compared with WT mice, mutants also showed a reduction in the percentage of sub-G0/G1 LSK cells, suggesting decreased apoptosis in this compartment. Compared with BM of mutant mice, larger proportion of LSK cells was quiescent in the mutant spleen. Collectively, these data suggest that in diseased mice, cells within the LSK compartment are driven out of quiescence, migrate out of the mutant BM, and colonize and expand in the splenic environment. Consistent with the increased number of LSK cells and the increase in cycling cells within this compartment, LSK cells from Ptpn11D61Y mice were hypersensitive to SCF and formed larger colonies compared with those from control (Figure S2 and data not shown).
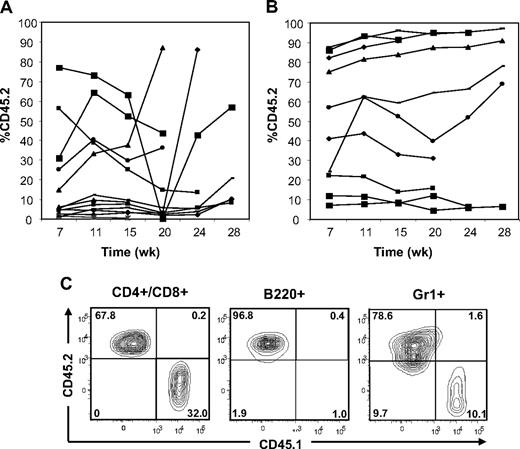
To ask whether adoptive transfer of BM or spleen cells from Ptpn11D61Y mice can confer MPD to WT recipients and gain initial insights into the effects of leukemogenic Ptpn11 on stem cell function, we performed BMT experiments. We injected 2 × 106 BM cells from diseased (> 6 months after pIpC induction) Ptpn11D61Y mice (donor) together with 105 CD45.1 WT BM cells for radioprotection into lethally irradiated, syngeneic (CD45.1) recipients. This corresponds to a 3-fold excess of phenotypic LT-HSCs from diseased mice. White blood cells (WBCs) and hematocrit (HCT) of recipients were monitored every 4 weeks. Lin− BM cells from Ptpn11D61Y mice showed equivalent homing efficiency to those of control mice (Figure S3A). At 7 weeks after transplantation, approximately half of the recipients showed a substantial percentage of donor-derived cells in their peripheral blood; however, these cells disappeared 15 to 20 weeks after transplantation (Figure 4A). Thereafter, some recipients of Ptpn11D61Y BM cells became moribund with a T-ALL/lymphoma-like syndrome, characterized by elevated WBCs and splenomegaly (Table 4 and data not shown). Others died, most likely of the same syndrome, although cells and tissue could not be recovered for detailed analysis. At no time did any of the recipients show progressive granulocytosis/monocytosis or other signs of MPD, nor did any show long-term multilineage reconstitution by donor cells (Table 4 and data not shown). Both BM and spleen cells from these recipients expressed Ptpn11D61Y RNA (Figure S3B). Based on these data, it appears that Ptpn11D61Y expression compromises BM stem cell number and/or function.
Adoptive transfer of BM cells and spleen cells from Ptpn11D61Y mice with MPD. (A) BM cells (2 × 106) or (B) spleen cells (5 × 106) from Ptpn11D61Y (CD45.2) mice were transferred together with 105 CD45.1 WT BM cells into lethally irradiated CD45.1 recipients. The percentage of peripheral blood cells expressing CD45.2 was quantified by flow cytometry at the indicated times. (C) Representative spleen cells from a recipient 24 weeks after transplantation, showing the presence of donor-derived B220+, CD3+/CD4+/CD8+, and Gr1+ cells.
Adoptive transfer of BM cells and spleen cells from Ptpn11D61Y mice with MPD. (A) BM cells (2 × 106) or (B) spleen cells (5 × 106) from Ptpn11D61Y (CD45.2) mice were transferred together with 105 CD45.1 WT BM cells into lethally irradiated CD45.1 recipients. The percentage of peripheral blood cells expressing CD45.2 was quantified by flow cytometry at the indicated times. (C) Representative spleen cells from a recipient 24 weeks after transplantation, showing the presence of donor-derived B220+, CD3+/CD4+/CD8+, and Gr1+ cells.
We also injected 5 × 106 splenocytes from Ptpn11D61Y mice along with 105 CD45.1 WT BM cells. This represents an even greater excess of phenotypic LT-HSCs (∼ 10-fold) from diseased mice. Unlike recipients of Ptpn11D61Y BM cells, approximately 30% to 40% of the recipients of mutant spleen cells showed stable engraftment with donor cells for more than 28 weeks after transplantation (Figure 4B). Five recipients died between 20 to 35 weeks of unknown causes, 1 developed donor (CD45.2)–derived T-ALL, and 2 developed recipient (CD45.1)–derived T-ALL (Figure S3C-D; Table 4; and data not shown). Again, none of the recipients that died showed progressive granulocytosis/monocytosis before their demise. Moreover, there was no evidence of MPD in recipients of Ptpn11D61Y spleen cells, even those with high donor chimerism (55%-95%). We performed necropsies on one such mouse at 25 weeks and another at 9 months; neither showed gross or histopathologic evidence of myeloid or lymphoid disease. In contrast, all primary Ptpn11D61Y animals tested showed elevated WBCs and overt splenomegaly at these time points (Figure 1 and data not shown). At 24 weeks after transplantation, surviving spleen cell recipients with high CD45.2 chimerism showed donor (Ptpn11D61Y)–derived multilineage reconstitution, including the aforementioned 2 “normal” mice subjected to detailed anatomic/histopathologic analysis (Figure 4C). PB cells from all surviving mice with high donor-derived chimerism were genotyped by polymerase chain reaction (PCR); all showed complete deletion of the STOP cassette, indicative of Ptpn11D61Y expression (Figure S3E and data not shown). We also sampled some of these recipients and found that they expressed Ptpn11D61Y RNA (Figure S3B). Together, these results suggest that spleens from mice with Ptpn11-evoked MPD contain functional HSCs. However, long-term stable engraftment of these mutant HSCs appears to be insufficient to give rise to MPD in irradiated wild-type recipients.
Cell-autonomous effects of Ptpn11D61Y on myeloid progenitors
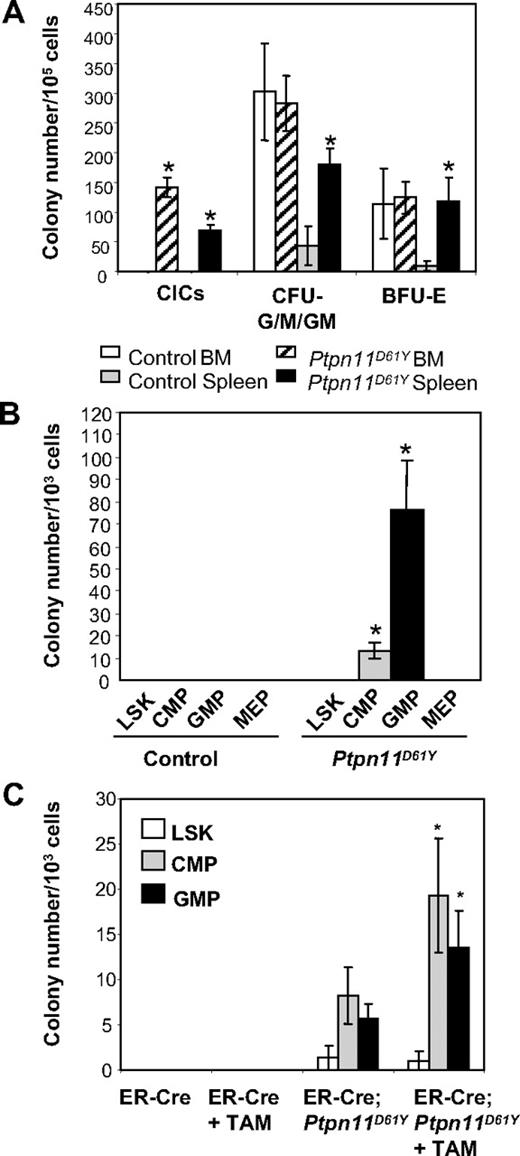
Diseased Ptpn11D61Y mice also had abnormalities in their committed progenitor compartments (as defined by surface phenotype). The proportion and absolute numbers of CMPs and GMPs were reduced in the BM of diseased mice (Table 2). In contrast, there were increases in the percentage and the absolute number of GMPs and the absolute number of MEPs and Ter119+ progenitors in the mutant spleens. Ptpn11D61Y spleen cells produced increased numbers of myeloid (CFU-G/M/GM) and erythroid (BFU-E) colonies under saturating doses of cytokines (Figure 5A). Mutant BM cells also produce more colonies in response to limited dose of either GM-CSF or M-CSF (Figure S4). Furthermore, in the absence of exogenous cytokines, both BM and spleen cells from diseased mice formed significant numbers of cytokine-independent colonies (CICs), which were predominantly macrophage colony-forming units (CFU-Ms) and granulocyte colony-forming units (CFU-Gs) (Figure 5A and data not shown).
Committed progenitors from Ptpn11D61Y mice evoke CICs. (A) Ptpn11D61Y and control BM and spleen cells were plated in methylcellulose media in the presence or absence of cytokines. (B) LSK cells, CMPs, GMPs, and MEPs from Ptpn11D61Y and control mice were purified by FACS and plated in methylcellulose media in the absence of cytokines. (C) LSK cells, CMPs, and GMPs from ER-Cre and ER-Cre;Ptpn11D61Y mice were purified and cultured for 16 hours in media containing SCF, LIF, IL-6, sIL-6R (for LSK), and SCF and IL-11 (for CMPs and GMPs) in the presence or the absence of Tam. Live cells were replated on methylcellulose medium in the absence of cytokine. For all experiments, colonies were counted after 7 to 9 days.
Committed progenitors from Ptpn11D61Y mice evoke CICs. (A) Ptpn11D61Y and control BM and spleen cells were plated in methylcellulose media in the presence or absence of cytokines. (B) LSK cells, CMPs, GMPs, and MEPs from Ptpn11D61Y and control mice were purified by FACS and plated in methylcellulose media in the absence of cytokines. (C) LSK cells, CMPs, and GMPs from ER-Cre and ER-Cre;Ptpn11D61Y mice were purified and cultured for 16 hours in media containing SCF, LIF, IL-6, sIL-6R (for LSK), and SCF and IL-11 (for CMPs and GMPs) in the presence or the absence of Tam. Live cells were replated on methylcellulose medium in the absence of cytokine. For all experiments, colonies were counted after 7 to 9 days.
To determine whether phenotypic HSCs and/or committed progenitors give rise to CICs, we purified LSK cells, CMPs, GMPs, and MEPs from Ptpn11D61Y mice and control littermates by fluorescence-activated cell sorting (FACS), and placed them in methylcellulose media in the absence of added growth factors. Only GMPs, and to a lesser extent, CMPs, gave rise to CICs, which again were either CFU-Ms or CFU-Gs (Figure 5B and data not shown). Similar results were obtained using purified progenitors from preleukemic animals in which Ptpn11D61Y expression had been induced for only a short (2-week) time (Figure S5A).
Although these data suggest that mutant committed progenitors form CICs, expression of Ptpn11D61Y might perturb the expression of the markers used to define these progenitor compartments. To exclude this possibility and to ask whether Ptpn11D61Y can directly confer factor independence on committed progenitors, we used BM from mice that coexpress the LSL-Ptpn11D61Y allele and a tamoxifen-inducible Cre (ER-Cre) transgene.19 LSK cells, CMPs, and GMPs were purified from ER-Cre and ER-Cre;LSL-Ptpn11D61Y mice, subjected to brief (16-hour) culture in the presence or absence of 4-hydroxytamoxifen (Tam), and then replated in cytokine-free methylcellulose medium. In the absence of Tam, ER-Cre;LSL-Ptpn11D61Y CMPs and GMPs gave rise to some CICs (Figure 5C), which correlated with modest Tam-independent in vivo deletion of the STOP cassette in ER-Cre;LSL-Ptpn11D61Y mice (data not shown). However, Tam-treated mutant CMPs and GMPs showed increased numbers of CICs compared with nontreated cells. In contrast, mutant LSK cells generated insignificant numbers of CICs in the absence of Tam, and did not show enhanced colony formation in the presence of Tam. Neither untreated nor Tam-treated LSK cells, CMPs, or GMPs harvested from ER-Cre mice produced any CICs (Figure 5C). These data establish that Ptpn11D61Y directly confers factor independence on myeloid progenitors in a cell-autonomous manner.
We asked whether CICs are driven by an autocrine mechanism. The levels of IL-3, GM-CSF, and M-CSF RNA in GMPs were quantified by quantitative polymerase chain reaction (qPCR). Neither control nor mutant GMPs produced significant amounts of IL-3 or GM-CSF; both produced similar amount of M-CSF (Figure S5B). Of course, we cannot exclude the possibility of autocrine production of other cytokines.
We next determined whether Ptpn11D61Y expression affects the differentiation potential of myeloid progenitors. For these experiments, CMPs and GMPs from control and ER-Cre;LSL-Ptpn11D61Y mice were plated in methylcellulose media with saturating doses of SCF, IL-3, IL-6, and EPO in the presence or absence of Tam. Control and mutant CMPs gave similar numbers of BFU-E, CFU-GEMM, and CFU-G/M/GM colonies in untreated cultures, and Tam addition did not affect the ratio of these colonies (Figure S5C). Similarly, GMPs from control and ER-Cre;LSL-Ptpn11D61Y mice produced comparable numbers of CFU-G/M/GM colonies in the absence and the presence of Tam (Figure S5C). Together, these data suggest that expression of Ptpn11D61Y does not alter the differentiation potential of CMPs and GMPs under saturating doses of cytokine. However, under in vitro conditions where growth factors are limiting, they give rise to myeloid cells with a monocytic bias.
Altered differentiation in Ptpn11D61Y erythroid progenitors
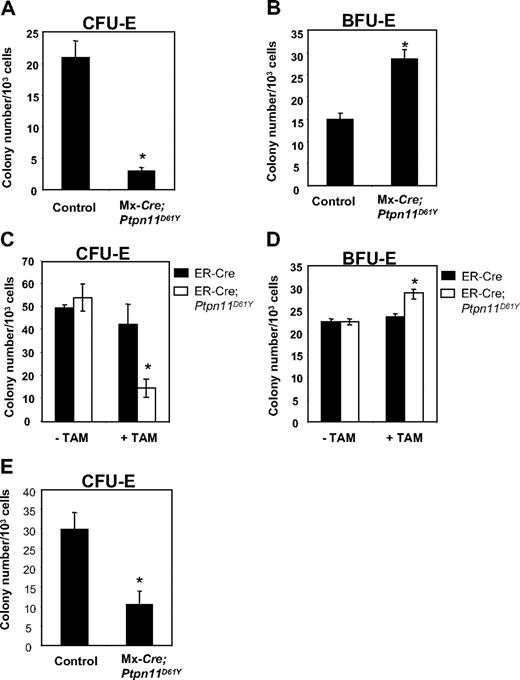
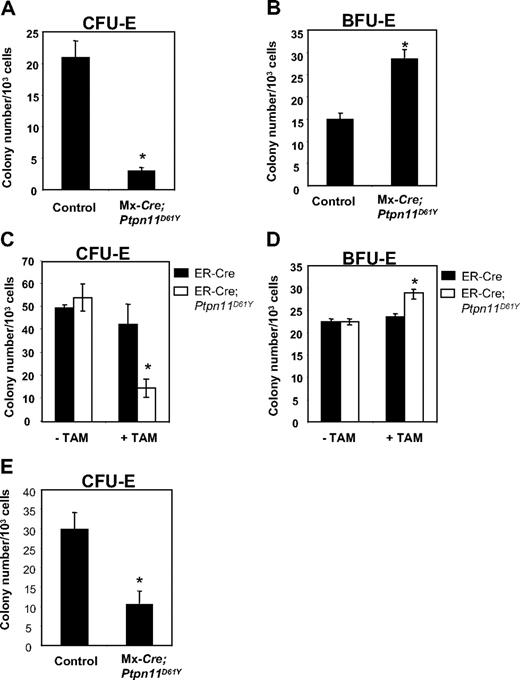
Ptpn11D61Y mice developed anemia, yet they showed a marked increase in splenic Ter119+ progenitors. Although the anemia could, at least in part, be due to hypersplenism, we asked whether erythroid differentiation was perturbed in mice with Ptpn11D16Y-evoked MPD. MEPs from BM of control and diseased Ptpn11D61Y mice were plated in methycellulose media containing EPO, and CFU-E and BFU-E colonies were enumerated after 2 or 8 days, respectively. Ptpn11D61Y MEPs consistently produced decreased numbers of CFU-E colonies (Figure 6A), while generating more (Figure 6B) and larger (data not shown) BFU-E colonies.
Ptpn11D61Y erythroid progenitors show enhanced BFU-E and reduced CFU-E activity. MEPs from control and Ptpn11D61Y (A,B) or ER-Cre;Ptpn11D61Y mice (C,D) were cultured for 2 days in EPO-containing methylcellulose media for CFU-E colonies (A,C) or for 8 days in medium containing SCF, IL-3, IL-6, and EPO for BFU-E colonies (B,D). (E) EPs from control and Ptpn11D61Y mice were plated on methylcellulose media in the presence of 0.5 U/mL EPO for 2 days before they were numerated.
Ptpn11D61Y erythroid progenitors show enhanced BFU-E and reduced CFU-E activity. MEPs from control and Ptpn11D61Y (A,B) or ER-Cre;Ptpn11D61Y mice (C,D) were cultured for 2 days in EPO-containing methylcellulose media for CFU-E colonies (A,C) or for 8 days in medium containing SCF, IL-3, IL-6, and EPO for BFU-E colonies (B,D). (E) EPs from control and Ptpn11D61Y mice were plated on methylcellulose media in the presence of 0.5 U/mL EPO for 2 days before they were numerated.
To determine whether these effects were cell autonomous and to rule out potential indirect consequences of excessive myelopoiesis on erythroid differentiation, we purified MEPs from ER-Cre and ER-Cre;LSL-Ptpn11D61Y mice and placed them in methylcellulose cultures with or without Tam. In the absence of Tam, we observed no difference in colony formation by control and mutant MEPs (Figure 6C,D). When Tam was present, however, MEPs from ER-Cre;LSL-Ptpn11D61Y BM produced substantially fewer CFU-E and slightly more BFU-E colonies compared with treated control cells (Figure 6C,D). Thus, Ptpn11D61Y, acting cell autonomously, alters the differentiation potential of erythroid progenitors within the MEP compartment.
Erythroid committed progenitors (EPs, Lin−cKit+Sca1−IL-7Rα−IL-3Rα−Ter119−CD41−CD71+) account for nearly all CFU-E activity in normal BM.20 EPs copurify with MEPs in WT20 and Ptpn11D61Y (data not shown) BM. We purified and plated equal numbers of EPs from control and mutant mice into EPO-containing methylcellulose medium. Similar to MEPs, Ptpn11D61Y EPs produced fewer CFU-E colonies compared with control cells (Figure 6E). These data suggest that the partial erythroid differentiation blockade caused by Ptpn11D61Y occurs at or distal to the EP stage.
Perturbed signaling in Ptpn11D61Y stem/progenitor cells
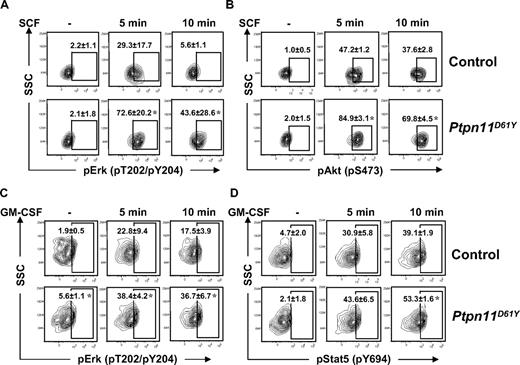
The effects of Ptpn11D61Y on different hematopoietic cell compartments described in previous sections suggested that mutant expression might have distinct, cell type–dependent consequences on growth factor/cytokine signaling. Recently, we developed a robust protocol for flow cytometric analysis of signaling proteins in HSCs and progenitor cells.21 Using this assay, we found that SCF-evoked Erk and Akt activation was substantially enhanced in LSK (Figure 7A,B) and LK (Figure S6A,B) cells from Ptpn11D61Y mice. There was no detectable activation of Stat3 or Stat5 in response to SCF in either WT or mutant LSK and LK cells (data not shown). In contrast, basal and GM-CSF–evoked Erk activation, as well as GM-CSF–evoked Stat5 activation, were enhanced in mutant CMPs and GMPs (Figure 7C,D, Figure S6C,D). We did not detect basal or GM-CSF–evoked Akt activation in WT or mutant progenitors (data not shown). Importantly, expression of the SCF receptor, Kit, on LSK and LK cells (Figure 3A), and GM-CSFR βc expression (data not shown) on GMPs and CMPs, as determined by flow cytometry, were unaffected by Ptpn11D61Y. Expression of GM-CSFRα could not be assessed due to lack of available antibodies for flow cytometry. Together, these data demonstrate that mutant LSK cells, GMPs, and CMPs show distinct and cell-intrinsic alternations in growth factor and cytokine responsiveness.
Signaling aberrations in Ptpn11D61Y cells. (A,B) Lin− BM cells from control and Ptpn11D61Y mice were purified by FACS and starved for 1 hour in serum-free medium before they were either left untreated or stimulated with 50 ng/mL SCF for 5 or 10 minutes. Cells were fixed, permeabilized, and stained with anti-cKit and anti-Sca1 and antibodies against pErk (A) and pAkt (B). Levels of phospho-specific antigens in the LSK population were determined by flow cytometry. (C,D) GMPs from control and Ptpn11D61Y mice were purified and starved before they were either left untreated or stimulated with GM-CSF (5 ng/mL) for 5 or 10 minutes. Cells were fixed, permeabilized, and stained with antibodies against pErk (C) and pStat5 (D), and levels of phospho-specific antigens quantified by flow cytometry (± SEM; *P < .05, by Wilcoxon-Mann-Whitney test).
Signaling aberrations in Ptpn11D61Y cells. (A,B) Lin− BM cells from control and Ptpn11D61Y mice were purified by FACS and starved for 1 hour in serum-free medium before they were either left untreated or stimulated with 50 ng/mL SCF for 5 or 10 minutes. Cells were fixed, permeabilized, and stained with anti-cKit and anti-Sca1 and antibodies against pErk (A) and pAkt (B). Levels of phospho-specific antigens in the LSK population were determined by flow cytometry. (C,D) GMPs from control and Ptpn11D61Y mice were purified and starved before they were either left untreated or stimulated with GM-CSF (5 ng/mL) for 5 or 10 minutes. Cells were fixed, permeabilized, and stained with antibodies against pErk (C) and pStat5 (D), and levels of phospho-specific antigens quantified by flow cytometry (± SEM; *P < .05, by Wilcoxon-Mann-Whitney test).
Discussion
We have established a new, inducible knockin model of Ptpn11-evoked MPD. As in our retroviral model,12 Ptpn11D61Y mice show myeloid expansion, elevated levels of myeloid precursors in the spleen, and tissue infiltration with granulocytes and macrophages. However, unlike the former model, in which only approximately 60% of mice develop myeloid disease with the remainder succumbing to T-ALL/lymphoma, MPD is fully penetrant in our inducible knockin mice, and nearly all Ptpn11D61Y mice develop anemia, a hallmark feature of JMML. Our initial characterization of Ptpn11D61Y mice has uncovered similarities to, and differences from, MPD caused by activating Kras and homozygous loss-of-function in Nf1 mutations.17,22,23
We find that leukemogenic Ptpn11 has multiple and distinct effects on different hematopoietic cell compartments, which collude to cause MPD (Figure S7). First, Ptpn11D61Y mice have markedly increased numbers of LSK cells. However, there is a “rightward” shift in this compartment, with a relative decrease in phenotypic LT-HSCs and increased percentages of phenotypic ST-HSCs and MPPs. The majority of mutant LSK cells are found in the spleen instead of the BM. Indeed, quiescent LSK cells are depleted in the BM of mutant mice, concomitant with an increase in cycling within the LSK compartment. There is a larger fraction of quiescent LSK cells within the spleen (compared with the BM) of mutant mice, although the percentage of cycling LSK cells there is also greater than in normal BM.
Yet although the LSK compartment is expanded, Ptpn11D61Y mice have decreased stem cell activity. We did not observe any long-term multilineage reconstitution after transplantation of Ptpn11D61Y BM cells, even though there was a 3-fold excess (over competitors) of phenotypic Ptpn11D61Y LT-HSCs. Likewise, only approximately 30% to 40% of mice that received a transplant of Ptpn11D61Y spleen cells (estimated to contain a 10-fold excess of phenotypic LT-HSCs compared with cotransplanted WT BM cells) show long-term reconstitution with donor cells. The fidelity of surface markers for specific stem and progenitor cell populations can be unreliable in mutant mice with aberrant hematopoiesis,24 so further studies will be required to determine whether stem cell number and/or stem cell activity is/are compromised in Ptpn11D61Y BM and spleen.
Some mice that received a transplant of Ptpn11D61Y spleen cells did show long-term multilineage reconstitution with Ptpn11D61Y-expressing cells, yet surprisingly, these recipients also failed to develop MPD. BM cells from mice with MPD evoked by retroviral expression of PTPN11 mutants also do not cause myeloid disease in secondary recipients.12 Furthermore, sublethally irradiated recipients engrafted with BM cells from mice with Nf1−/− MPD fail to develop disease, even though all their hematopoietic cells are donor derived.22 In contrast, the MPD that develops in KrasG12D and Nf1−/− mice reportedly can be transplanted into lethally irradiated recipients.22,23
There are several possible explanations for the failure to transplant Ptpn11D61Y-evoked MPD. First, the MPD-initiating stem cell could be rare (Figure S8). If so, this could reflect the need for a second hit (besides Ptpn11D61Y expression) for disease initiation. Alternatively, MPD may be evoked only by the expression of Ptpn11D61Y in a relatively rare subpopulation of HSCs. For example, Ptpn11D61Y might have to target a juvenile, or even a remnant fetal HSC, a potentially attractive model that might explain why NS patients are at risk of JMML only at a young age.1 Consistent with this “rare cell” model, a critical threshold of donor-derived mutant cells is required to initiate MPD in the Nf1−/− mouse model.25 The second major alternative is that the niche of recipient mice does not support MPD. For example, lethal irradiation could result in a hostile environment for the engraftment of the Ptpn11D61Y MPD stem cell. Alternatively, a fetal/juvenile, rather than an adult, niche may be required. Conceivably, the oncogenic allele must be expressed in stromal cells as well as the hematopoietic compartment for MPD development.26,27 However, this seems unlikely for Ptpn11D61Y-evoked MPD, given that WT mice receiving BM cells transduced with mutant Ptpn11 alleles develop myeloid disease with a similar severity and kinetics as that of the induced Mx1-Cre-Ptpn11D61Y mice.12,28 Moreover, in sporadic JMML, the oncogenic allele presumably is restricted to the hematopoietic compartment, not the stroma.
Further research will be required to distinguish between these models. We also cannot exclude the possibility that competitor cells that were cotransplanted with mutant cells compete for the stem cell niche and suppress MPD development. However, this seems unlikely for those recipients that showed high donor chimerism, yet did not develop disease. Our findings, along with the apparent impairment of stem cell activity in Ptpn11D61Y mice, raise the possibility that mutant HSCs lose self-renewal activity during disease progression. This might explain the spontaneous hematologic improvement seen in some rare JMML patients,1 and suggests that it may be possible to “burn-out” MPD-initiating cells in others.
Although the expanded LSK compartment probably plays an important role in the development of Ptpn11-evoked MPD (most likely by delivering excess cells to the committed progenitor compartment), merely increasing LSK numbers probably is not sufficient to cause MPD when distal homeostatic regulation is intact.29,30 Similar to earlier retroviral studies,10-12 Ptpn11D61Y BM and spleen cells produce myeloid colonies in the absence of exogenous cytokines. Our studies show that these colonies arise from early committed myeloid progenitors (CMPs and GMPs), not the LSK compartment. The ability of myeloid progenitors to survive/proliferate/differentiate under limited cytokine conditions may render these precursors unable to respond to normal homeostatic regulation and explain the selective expansion of the myeloid lineage when Ptpn11D61Y is ubiquitously expressed in all hematopoietic cells. This, together with the excess progenitors produced by the expanded LSK compartment, probably accounts for myelomonocytosis in these mice (Figure S7).
In contrast, Ptpn11D61Y has distinct effects on different stages of erythropoiesis, which ultimately results in anemia. It was reported previously that BM and fetal liver (FL) cells infected with an activated allele of Ptpn11 form increased numbers of, and larger, BFU-E colonies.11 Consistent with that report, Ptpn11D61Y MEPs produce increased numbers of, and larger, BFU-E colonies. This suggests increased conversion of bipotential cells within the MEP compartment to the erythroid lineage, as well as increased proliferation of early-stage erythroid cells. Consistent with the latter conclusion, MEP-enriched CD34−LK cells from Ptpn11D61Y mice show enhanced Erk and Akt activation after stimulation by SCF (data not shown), a cytokine required for BFU-E colony formation. In contrast, Ptpn11D61Y MEPs, in a cell-autonomous manner, produce fewer CFU-E colonies, suggesting a partial block in erythroid differentiation/survival at the CFU-E stage or later. Initial experiments indicate that selective expression of Ptpn11D61Y in the erythroid lineage using EPOR-Cre31 also results in anemia (n = 5, data not shown), and MEPs from these mutant mice also produce more BFU-E and fewer CFU-E colonies (n = 2, data not shown).
Exactly where in the erythroid differentiation pathway this defect occurs remains to be determined. Ptpn11D61Y EPs also produce fewer CFU-E colonies; hence, defective CFU-E colony production by mutant EPs may account for most, if not all, of the reduced CFU-E activity in the MEP compartment. Notably, FL cells from KrasG12D mice32,33 also show a partial block at the CD71hiTer119−/low progenitor stage, which contains CFU-Es.34
Excess proliferation and transformation require subversion of normal signaling pathways. We found that LSK and LK cells from Ptpn11D61Y mice are hypersensitive to SCF, and show enhanced phosphorylation of Erk, Akt, and S6 (Figure 7, S6 and data not shown) after SCF stimulation. In contrast, SCF-evoked phosphorylation of Erk and S6 are attenuated in LK cells from KrasG12D mice.35 These differences are surprising given that Kras and Shp2 act on the Ras/Erk pathway, and KrasG12D and Ptpn11D61Y mice develop grossly similar MPDs.17,23 Although these discrepancies could reflect technical differences, our data suggest that different disease alleles evoke distinct biochemical profiles in the same pathway in the same cell type, differences that may have therapeutic implications.
Myeloid colony growth in the absence of exogenous stimulation and the selective hypersensitivity of BM cells to GM-CSF are hallmarks of JMML.36,37 GM-CSF signaling is also required for fully penetrant MPD in Nf1−/− mice38,39 and the GM-CSF analog, E21R, inhibits JMML cell proliferation in vitro, in a mouse xenograft model and transiently in a JMML patient.1 We find that GMPs and CMPs, upon acute expression of Ptpn11D61Y in vitro and in vivo, give rise to myeloid colonies in the absence of exogenous cytokines. Furthermore, these mutant progenitors show higher basal phosphorylation of Erk and S6, and enhanced phosphorylation of Erk, Stat5, and S6 after GM-CSF stimulation (Figures 7, S6, and data not shown), which is similar to effects reported in LK cells from KrasG12D mice.35 As both KrasG12D and Ptpn11D61Y cells show elevated GM-CSF–induced Stat5 phosphorylation, future experiments should determine whether this is a result of enhanced activation of Jak2 and assess the potential efficacy of the novel Jak2 inhibitors40,41 in treatment of MPD in these animals. Interestingly, a recent report shows that BM cells of JMML patients evoke a unique STAT5 activation signature after GM-CSF stimulation,42 further supporting the therapeutic evaluation of Jak2 inhibitors. Our Ptpn11D61Y model, together with KrasG12D and Nf1−/− mice, should facilitate the evaluation of these and other promising therapeutic strategies, as well as provide more insights in the molecular basis for leukemia initiated by abnormal activation of the Ras/Erk pathway.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Norman Iscove (Ontario Cancer Institute [OCI]) for critical reading of this paper; John Daley (Dana-Farber Cancer Institute [DFCI]), Susan Lazo-Kallanian (DFCI), Michelle Tseng (Hospital for Sick Children, [HSC], Toronto, ON) Leanne Jamieson (HSC), and Pier-André Pentillä (HSC) for expert assistance with flow cytometry; and Jason Gilliland (Beth Israel Deaconess Medical Center, [BIDMC]) Sonja Boet-Whitaker (BIDMC), Tarun Sharma (OCI), and Ashley Sanders (OCI) for technical assistance.
D.K. was supported by a National Institutes of Health (NIH; Bethesda, MD) postdoctoral institutional training grant in hematology (5T32-HL07623-20). M.G.M. was supported by a fellowship from the Deutsche José Carreras Leukämie-Stiftung e.V. (München, Germany; and a scholar award from the American Society of Hematology (Washington, DC). This work was supported by NIH RO1 CA114945 awarded to B.G.N.
National Institutes of Health
Authorship
Contribution: G.C. designed research, performed research, analyzed data, and wrote the paper; D.K. and T.U. designed research, performed research, and analyzed data; J.L.K. performed research and analyzed data; W.T. and M.G.M performed research; and B.G.N. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current address for Dr Kalaitzidis is Division of Hematology, Brigham and Women's Hospital, Boston, MA. The current address for Dr Yang is Department of Orthopedics, Brown University Alpert Medical School, Providence, RI. The current address for Dr Mohi is Department of Pharmacology, State University of New York (SUNY) Upstate Medical University, Syracuse, NY.
Correspondence: Gordon Chan, Department of Stem Cell and Developmental Biology, Ontario Cancer Institute, 101 College St, TMDT Rm 8-301, Toronto, ON M5G 1L7, Canada; e-mail: gordon.chan@uhnresearch.ca.