Abstract
We investigated human leukocyte antigen (HLA) expression on leukemic cells derived from patients at diagnosis and relapse after hematopoietic stem cell transplantation (HSCT) using flow cytometry with locus-specific antibodies. Two of 3 patients who relapsed after HLA-haploidentical HSCT demonstrated loss of HLA alleles in leukemic cells at relapse; on the other hand, no loss of HLA alleles was seen in 6 patients who relapsed after HLA-identical HSCT. Single-nucleotide polymorphism array analyses of sorted leukemic cells further revealed the copy number-neutral loss of heterozygosity, namely, acquired uniparental disomy on the short arm of chromosome 6, resulting in the total loss of the mismatched HLA haplotype. These results suggest that the escape from immunosurveillance by the loss of mismatched HLA alleles may be a crucial mechanism of relapse after HLA-haploidentical HSCT. Accordingly, the status of mismatched HLA on relapsed leukemic cells should be checked before donor lymphocyte infusion.
Introduction
Human leukocyte antigen (HLA) molecules expressed on the cell surface are required in presenting antigens to T cells. The HLA class I antigens are vital in the recognition of tumor cells by tumor-specific cytotoxic T cells. The loss of HLA class I molecules on the cell surface membrane may lead to escape from T-cell immunosurveillance and the relapse of leukemia. Previously, loss of HLA class I haplotype has been described in solid tumors.1-3 However, there are few reports concerning HLA-haplotype loss in leukemia.4,5
We examined HLA class I expression in leukemic blasts from patients who relapsed after hematopoietic stem cell transplantation (HSCT) to analyze whether the loss of HLA on leukemic cells was related to the relapse after HLA-identical or haploidentical HSCT.
Methods
Patients and transplantation procedure
We identified 9 children with acute leukemia who relapsed after HSCT. Their leukemic samples were cryopreserved both at the time of the initial diagnosis and of relapse. The patients' characteristics are summarized in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Three patients received HSCT from an HLA-haploidentical family donor, and the other 6 patients received HSCT from an HLA-matched donor (4 siblings and 2 unrelated donors).
Written informed consent was given by the parents according to the protocol approved by the ethics committee of Nagoya University Graduate School of Medicine in accordance with the Declaration of Helsinki.
HLA class I expression on leukemic cells
Samples were collected at diagnosis and post-transplantation relapse. HLA expression of leukemic blasts and normal cells was analyzed by flow cytometry as previously reported.6 Anti-HLA A2-FITC (cloneBB7.2) and anti-HLA A24-FITC (clone17a10) monoclonal antibodies were purchased from Medical & Biological Laboratories; HLA-A11 (IgM), HLA-A30, HLA-31 (IgM), HLA-25, HLA-26 (IgM), HLA-Bw6 (IgG3), and HLA-Bw4 (IgG3) antibodies were purchased from One Lambda. For leukemic cell markers, CD13-PE (IgG1) were purchased from Immunotech and CD34-APC (IgG1) were purchased from BD Biosciences. Samples were analyzed with FACSCalibur cytometer and CellQuest software. The method of genomic HLA typing was previously reported.7
Isolation of DNA and single nucleotide polymorphism analysis
The CD13+/CD34+ leukemic blasts were sorted by flow cytometry from bone marrow cells at the time of diagnosis and of relapse. Genomic DNA was extracted from leukemic cells sorted by a fluorescence-activated cell sorter as well as from phytohemagglutinin-stimulated patient-derived T cells and subjected to single nucleotide polymorphism (SNP) array analysis using GeneChip NspI arrays (Affymetrix) according to the manufacturer's protocol. Allele-specific copy number was detected using Copy Number Analyzer for GeneChip software as previously described.8
Limiting dilution-based CTLp frequency assay
The frequencies of cytotoxic T-lymphocyte precursor (CTLp) specific for the recipient-mismatched HLA molecules were analyzed using a standard limiting dilution assay.9
Cytotoxic assay of CTL clones against leukemic blasts and a mismatched HLA cDNA-transfected B-lymphoblastoid cell line
The remaining cells of several cytotoxicity-positive wells used for the CTLp assay for the donor were used to obtain allo-HLA–restricted CTLs. CTL clones were isolated by standard limiting dilution and expanded as previously described.10,11
The HLA class I–deficient 721.221 B-lymphoblastoid cell line was maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2mM l-glutamine, and 1mM sodium pyruvate. Retroviral transduction was conducted as previously described.12
The cytotoxicity of CTL clones against target cells was analyzed by conventional chromium 51 (51Cr) release assay as previously reported.13
CTL clones (104 cells/well) were mixed with the indicated stimulator cells (104 cells/well) in 96-well, round-bottom polypropylene plates and spun at 1200g for 3 minutes before overnight incubation in 200 μL of RPMI 1640 medium supplemented with 10% fetal bovine serum. On the next day, 50 μL of supernatant was collected and interferon-γ was measured by enzyme-linked immunosorbent assay with 3,3′,5,5′-tetramethylbenzidine substrate (Sigma-Aldrich).
Results and discussion
Three children with high-risk acute myelogenous leukemia (AML) received haploidentical grafts from their parents but relapsed 8, 14, and 15 months after HSCT. Patient 2 received 3 courses of donor lymphocyte infusion (DLI) for relapsed leukemia after haploidentical HSCT. After the third unmanipulated DLI (107 CD3+/kg), she experienced acute grade-III graft-versus-host disease and achieved complete remission. However, she experienced a second relapse 6 months later. To monitor residual disease in those patients, we used flow cytometric analysis with antibodies specific for the mismatched HLA alleles between the donor and patient. Surprisingly, we found total loss of HLA-A2 expression on CD13+/CD34+ leukemic cells from bone marrow in 2 of 3 patients who underwent HLA-haploidentical HSCT, whereas microscopic analysis showed relapse (Figure 1A). To test whether HLA class I molecules could be up-regulated, samples were cultured for 48 hours in medium supplemented with tumor necrosis factor-α or interferon-γ and measured again; however, no restoration was observed (data not shown).
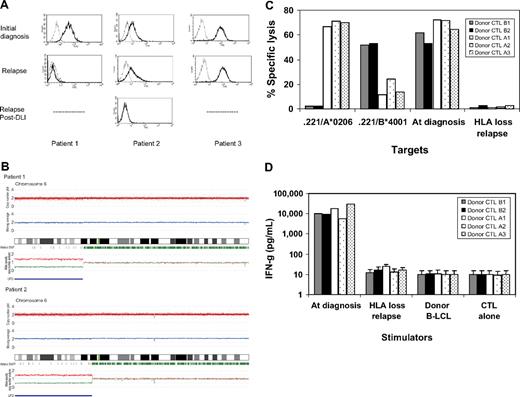
The loss of mismatched HLA expression on leukemic blasts caused by uniparental disomy on chromosome 6p impaired recognition and killing of donor's alloreactive cytotoxic T lymphocytes. (A) Leukemic blasts at the time of initial diagnosis and at the time of relapse after hematopoietic stem cell transplantation (HSCT) and donor lymphocyte infusion (DLI) were gated by CD34+ and CD13+, and then the surface expression of mismatched human leukocyte antigen (HLA) alleles was examined with anti–HLA-A2 antibodies. In 3 patients with acute myelogenous leukemia (AML) who experienced relapse after HLA-haploidentical HSCT, HLA-A2 expression was lost in patient 1 at relapse 15 months after HSCT and lost in patient 2 at second relapse 6 months after DLI. (B) Single nucleotide polymorphism (SNP) array analyses of sorted leukemic cells with the loss of an HLA allele revealed that the short arm of chromosome 6 shows copy number-neutral loss of heterozygosity or acquired uniparental disomy as detected by dissociated allele-specific copy number plots (red and blue lines at the bottom), resulting in the total loss of the mismatched HLA haplotype in both patient 1 and patient 2. The presence of acquired uniparental disomy is also indicated by normal total copy numbers with missing heterozygous SNPs (green bars) in the distal part of the short arm. (C) Recipient alloantigen-specific cytotoxic T-lymphocyte (CTL) clones were generated by a conventional cloning method from cytotoxicity-positive wells obtained in the limiting dilution assays using the donor CD8+ cells as responders. Donor CTL clones A1, A2, and A3 were specific for HLA-A*0206. Donor CTL clones B1 and B3 were specific for HLA-B*4001, all of which recognize mismatched HLA alleles between the donor and recipient. Those 5 representative CTL clones were tested for HLA specificity and recognition of leukemic blasts obtained at the time of the initial diagnosis and at the time of HLA loss relapse after DLI by a standard 51Cr-release assay at the effector/target ratio of 30:1. (D) Their interferon-γ production was also assessed against leukemic blasts collected at the time of diagnosis and at the time of HLA-loss relapse.
The loss of mismatched HLA expression on leukemic blasts caused by uniparental disomy on chromosome 6p impaired recognition and killing of donor's alloreactive cytotoxic T lymphocytes. (A) Leukemic blasts at the time of initial diagnosis and at the time of relapse after hematopoietic stem cell transplantation (HSCT) and donor lymphocyte infusion (DLI) were gated by CD34+ and CD13+, and then the surface expression of mismatched human leukocyte antigen (HLA) alleles was examined with anti–HLA-A2 antibodies. In 3 patients with acute myelogenous leukemia (AML) who experienced relapse after HLA-haploidentical HSCT, HLA-A2 expression was lost in patient 1 at relapse 15 months after HSCT and lost in patient 2 at second relapse 6 months after DLI. (B) Single nucleotide polymorphism (SNP) array analyses of sorted leukemic cells with the loss of an HLA allele revealed that the short arm of chromosome 6 shows copy number-neutral loss of heterozygosity or acquired uniparental disomy as detected by dissociated allele-specific copy number plots (red and blue lines at the bottom), resulting in the total loss of the mismatched HLA haplotype in both patient 1 and patient 2. The presence of acquired uniparental disomy is also indicated by normal total copy numbers with missing heterozygous SNPs (green bars) in the distal part of the short arm. (C) Recipient alloantigen-specific cytotoxic T-lymphocyte (CTL) clones were generated by a conventional cloning method from cytotoxicity-positive wells obtained in the limiting dilution assays using the donor CD8+ cells as responders. Donor CTL clones A1, A2, and A3 were specific for HLA-A*0206. Donor CTL clones B1 and B3 were specific for HLA-B*4001, all of which recognize mismatched HLA alleles between the donor and recipient. Those 5 representative CTL clones were tested for HLA specificity and recognition of leukemic blasts obtained at the time of the initial diagnosis and at the time of HLA loss relapse after DLI by a standard 51Cr-release assay at the effector/target ratio of 30:1. (D) Their interferon-γ production was also assessed against leukemic blasts collected at the time of diagnosis and at the time of HLA-loss relapse.
Next, to examine the potential loss of genes encoding the undetectable HLA alleles, we sorted CD13+/CD34+ leukemic blasts and performed DNA genotyping. We found that, in addition to the HLA-A locus, the HLA-B, -C, and -DR loci were not encoded; only the mismatched haplotype was lost in both patients (supplemental Table 2). We then questioned whether this phenomenon would also occur in HLA-matched HSCT settings using anti-HLA class I antibodies. We did not observe any loss of HLA class I expression in any of the patients at the time of relapse (supplemental Figure 1). These results suggest that loss of HLA class I haplotype at the time of posttransplantation relapse is uncommon in HLA-matched HSCT.
To elucidate the mechanism of the loss of the mismatched HLA haplotype, we performed an SNP array analysis of genomic DNA extracted from leukemic blasts at the time of diagnosis and of relapse. Genomic DNA from patient-derived T cells was used as a reference. Leukemic cells at the time of relapse showed copy number-neutral loss of heterozygosity or an acquired uniparental disomy (UPD) of the short arm of chromosome 6 encompassing the HLA locus, whereas no allelic imbalance was identified at the time of diagnosis (Figure 1B). Loss of one allele from one parent and duplication of the remaining allele from the other parent led to UPD.14
In patient 2, we examined whether the number of CTLp had changed during the posttransplantation course. Limiting dilution analysis with a split-well 51Cr-release assay was carried out to compare the CTLp frequencies specific for the mismatched antigens between the recipient and donor. Interestingly, the CTLp frequencies were recovered after DLI (Table 1). Restoration of CTLp after 3 DLIs could eradicate such leukemic cells, lasting for 6 months thereafter.
Next, we generated allo-HLA–restricted CTLs from CD8+ cells obtained at day 520 in patient 2 and tested with the 721.221 B-lymphoblastoid cell line transfected with 1 of 3 mismatched HLA alleles (Figure 1C-D).
Despite high transplantation-related mortality resulting from severe graft-versus-host disease and posttransplantation infections, haploidentical HSCT has been widely used with the expectation of a strong graft-versus-leukemia effect.15 However, our observation provides a possible limitation of this strategy. Indeed, 2 of 3 patients showed genomic loss of the recipient-specific HLA-haplotype, which led to escape from the graft-versus-leukemia effect and relapse of the disease.
Vago et al also reported a similar observation in 5 of 17 (29.4%) patients whose disease relapsed after haploidentical HSCT.16 Relapsed leukemic cells may possess genomic instability that elicits genetic diversity.17 Immunologic pressure by alloreaction to major HLA antigens may select leukemic variants of HLA class I loss, which results in the survival and proliferation of these variants.
In haploidentical HSCT, the importance of natural killer (NK)–cell alloreactivity is emphasized to achieve the graft-versus-leukemia effect.18,19 HLA loss on leukemic blasts may in turn enhance the NK-cell alloreactivity. Our 2 patients with HLA loss had a group 1 homozygous HLA-C locus that is a suppressive killer immunoglobulin-like receptor (KIR) for NK cells and a KIR-matched donor (supplemental Table 2). Because UPD does not change the total copy number of the gene, donor NK cells should have been suppressed even after UPD occurred in these patients. Interestingly, the remaining patient who experienced relapse without HLA loss after HLA-haploidentical HSCT had a KIR-mismatched donor, so alloreactive NK cells were possibly enhanced to kill leukemic blasts with HLA loss.
Although one limitation of our study is an insufficient number of cases, our results combined with those in a recent report16 suggest that leukemic cells occasionally escape from immunosurveillance through the loss of the mismatched HLA haplotype by the mechanism of UPD after haploidentical HSCT. DLI for relapsed AML is less effective than that for chronic myelogenous leukemia after HLA-matched HSCT.20 However, DLI is effective even for the relapse of AML after haploidentical HSCT.21 Evaluation of loss or down-regulation of HLA on relapsed leukemic blasts after HLA-haploidentical HSCT should be considered because DLI would probably be ineffective in patients whose leukemic cells lose HLA class I antigen.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the Ministry of Education, Culture, Science, Sports, and Technology, Japan (Grant for Scientific Research on Priority Areas; B01 no. 17016089); Grants for Research on the Human Genome, Tissue Engineering Food Biotechnology, and the Second and Third Team Comprehensive 10-year Strategy for Cancer Control (no. 26) from the Ministry of Health, Labor, and Welfare, Japan; Core Research for Evolutional Science and Technology of Japan (Grant-in-Aid); the College Women's Association of Japan (scholarship award, I.B.V.); a grant from Foundation for Promotion of Cancer Research; and a grant from Morinaga Hoshikai and Grant-in-Aid for Scientific Research (c) No. 20591252.
Authorship
Contribution: I.B.V. performed experiments and wrote the manuscript; Y.T. designed the research, analyzed data, and wrote the manuscript; Y.A., H.S., M.K., and S.O. performed experiments, analyzed data, and wrote the manuscript; S.K. supervised this work and wrote the manuscript; and all other authors were responsible for clinical work and critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Seiji Kojima, Department of Pediatrics, Nagoya University Graduate School of Medicine, 65 Tsurumai-cho, Showa-ku, Nagoya, 466-8550, Japan; e-mail: kojimas@med.nagoya-u.ac.jp.