Abstract
We report gene expression and other analyses to elucidate the molecular characteristics of acute lymphoblastic leukemia (ALL) in children with Down syndrome (DS). We find that by gene expression DS-ALL is a highly heterogeneous disease not definable as a unique entity. Nevertheless, 62% (33/53) of the DS-ALL samples analyzed were characterized by high expression of the type I cytokine receptor CRLF2 caused by either immunoglobulin heavy locus (IgH@) translocations or by interstitial deletions creating chimeric transcripts P2RY8-CRLF2. In 3 of these 33 patients, a novel activating somatic mutation, F232C in CRLF2, was identified. Consistent with our previous research, mutations in R683 of JAK2 were identified in 10 specimens (19% of the patients) and, interestingly, all 10 had high CRLF2 expression. Cytokine receptor-like factor 2 (CRLF2) and mutated Janus kinase 2 (Jak2) cooperated in conferring cytokine-independent growth to BaF3 pro-B cells. Intriguingly, the gene expression signature of DS-ALL is enriched with DNA damage and BCL6 responsive genes, suggesting the possibility of B-cell lymphocytic genomic instability. Thus, DS confers increased risk for genetically highly diverse ALLs with frequent overexpression of CRLF2, associated with activating mutations in the receptor itself or in JAK2. Our data also suggest that the majority of DS children with ALL may benefit from therapy blocking the CRLF2/JAK2 pathways.
Introduction
Children with Down syndrome (DS) have a higher rate of acute lymphoblastic leukemia (DS-ALL). DS-ALLs are mostly of B-cell precursor (BCP) origin and similar in the age of diagnosis and immunophenotype to high hyperdiploid (HD) or TEL-AML1 ALLs,1 the 2 most common genetic subtypes of childhood ALL. Given that these cytogenetic abnormalities are less frequent in DS-ALL,2 the existence of unique collaborating somatic genetic events in DS-ALL, similar to the GATA1 mutation in DS-acute megakaryoblastic leukemia,3 has been postulated.
We and others reported the presence of somatic activating mutations in JAK2 in approximately 20% of DS-ALLs.4-6 Similar mutations are present in approximately 10% of high-risk ALLs in non-DS children, corresponding to approximately 3% of unselected childhood ALLs.7 We hypothesized that the mutated Janus kinase 2 (JAK2) may cooperate with a type I cytokine receptor that is aberrantly expressed in DS-ALL.4
To characterize additional molecular abnormalities in DS-ALL, we performed genomic analysis of a large group of DS-ALLs. This analysis reveals, next to a striking heterogeneity of these leukemias, an aberrant expression of the cytokine receptor CRLF2 in 62% of the patients, associated with somatic activating mutations in JAK2 or in the receptor itself.
Methods
Patient samples
RNA and DNA were derived from diagnostic bone marrow samples of children with DS and BCP-ALL enrolled on treatment protocols with an informed consent and approval of the ethics committees of all participating institutions in accordance with the Declaration of Helsinki. Samples were anonymized for the study. Patients' clinical data are described in supplemental Tables 1 and 2 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Seventy-six of these patients were included in our previous publication describing the JAK2 mutations in DS-ALL.4 The study was approved by the Israeli Health Ministry Ethic committee (approval no. 920070771).
Genomic studies
RNA processing and hybridization to Affymetrix arrays were performed according to the manufacturer's instructions and as previously published.8,9 Only specimens containing more than 70% blasts were included. There were 4 datasets obtained by different teams as summarized in Table 1. Associazione Italiana Ematologia Oncologia Pediatrica (AIEOP) is the main dataset used for gene expression analysis, comprising 97 diagnostic ALL samples: 25 DS-ALL, and 72 non–DS-ALL samples, described in Table 1. The additional 3 datasets were used for validations. The primary gene expression data files have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/)11 (GEO series accession no. GSE17459).
Genomic DNA from 42 diagnostic bone marrow ALLs and 34 paired remission samples were genotyped with Affymetrix GeneChip Human Mapping 100K set (Affymetrix) according to the manufacturer's directions. See supplemental data for details.
Mutation analysis
Quantitative real-time PCR
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using Applied Biosystems TaqMan Gene-Expression Assays (CRLF2 Hs_00845692, GAPDH Hs_99999905) according to the manufacturer's instructions. Each sample was run in triplicate. The endogenous control gene was GAPDH.
FISH
Fluorescence in situ hybridization (FISH) for detection of IGH@-CRLF2 translocation or the presence of a microdeletion upstream to CRLF2 was performed as described.13
Flow cytometric analysis
Flow cytometric analysis (FACSCanto II [Becton Dickinson]; FlowJo software [TreeStar]) was performed on primary cryopreserved ALL cells after the first xenotransplantation in Nod/LtSzScid interleukin-2γ (IL2γ)–null mice. Antibodies used were anti–cytokine receptor-like factor 2 (CRLF2; clone 1D3, ab48482; Abcam), goat anti–mouse Alexa Fluor 488 (Invitrogen), anti-IL7RA Alexa 647 (CD127, clone HIL-7R-M21; BD), anti-CD19 phycoerythrin (clone LT19, MCA1940; AbD Serotec), and 7-amino-actinomycin D (AbD Serotec). All samples were gated on the viable (7-amino-actinomycin D–negative) and leukemic (CD19+) population before analysis of CRLF2 and IL7RA. For the calculation of delta mean fluorescence intensity (MFI), background nonspecific staining was evaluated in populations gated by CD19, comparing tubes with or without anti-CRLF2 antibodies. This background MFI was similar to the MFI of CRLF2-negative populations in normal human blood.
Plasmid construction
The FLAG-mJak2 wild-type and R683S were cloned into the pHRSINCSGW lentivirus,12 which carries spleen focus-forming virus (SFFV) promoter and an emerald green fluorescent protein reporter. pMX-Puro-hCRLF2 was used as a template for the generation of CRLF2 mutations by site-directed mutagenesis (QuikChange-II-XL; Stratagene).
Cell lines
BaF3 cells were cultured in RPMI-1640 containing 10% fetal calf serum and 10% WHEI-3B conditioned media as a source of interleukin-3.
Parental BaF3 cells were transduced with pMX-Puro-hCRLF2,14 and hCRLF2-expressing cells were selected with puromycin (2 μg/mL). Parental BaF3 and BaF3-CRLF2 cells were transduced with the appropriate Jak2-expressing vector and green fluorescent protein–positive cells were sorted by flow cytometry 3 to 4 days later.
BaF3 proliferation assays and Western blotting
BaF3 proliferation assays and Western blotting were performed as described before.4 Antibodies used were anti-JAK2 (C-20; Santa Cruz Biotechnology), anti–signal transducer and activator of transcription 5 (STAT5), anti–phospho-JAK2 Tyr1007 (Cell Signaling Technology), anti–phospho-STAT5 Tyr694 (Epitomics), anti–human thymic stromal lymphopoietin (hTSLPR; AF981; R&D Systems), anti–FLAG-M2, and anti–α-tubulin (Sigma-Aldrich).
Pharmacologic inhibition of JAK2
BaF3 cells expressing Jak2 R683S and BaF3/CRLF2 cells expressing wild-type (wt) or R683S Jak2 were cultured without cytokines in different concentrations of JAK inhibitor I (Calbiochem). Controls were BaF3/EpoR cells expressing BCR-ABL. Viable cells were counted after 72 hours. Data from 3 independent experiments were combined for analysis. We calculated the normalized viability by dividing the cell number at each inhibitor concentration by the cell number with vehicle alone.
Bioinformatics
Gene expression preprocessing is described in supplemental Methods.
Combining probe sets of the same gene.
For those genes that were represented by more than one probe set, we used, when needed, a combination procedure to create a single representation of a gene's expression (supplemental Methods).
GSEA.
The first ingredient of Gene Set Enrichment Analysis (GSEA)15 is a list of genes (L), ranked by some attribute (A), ordered from low to high values of A. The second ingredient is a set of genes (S) that is a subset of L. GSEA aims at answering whether the members of S are randomly distributed along the ranked list L, or whether they are skewed toward one of the sides. For details see supplemental Methods and Subramanian et al.15
Refining DS-ALL profile genes.
The preliminary DS-ALL profile gene list, which was constructed using the AIEOP dataset, was narrowed down using GSEA15 to select genes that show consistent expression pattern in at least 2 of the other 3 datasets (Table 1). We used the up-regulated members of the preliminary DS-ALL genes as our set S (see “Bioinformatics”) and the genes of 1 of the other 3 experiments constituted L. The genes of L were ordered according to their differential expression in DS versus the rest of the samples (see Refining DS-ALL profile genes in supplemental Methods). The process was repeated for each of the 3 datasets and for the down-regulated genes, yielding for each case genes that were identified as consistently up-regulated (or down-regulated) in the AIEOP dataset and the other dataset tested (Figure 2A).
Results
Marked heterogeneity of DS-ALLs revealed by unsupervised gene expression analysis
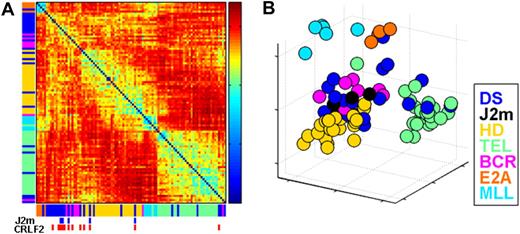
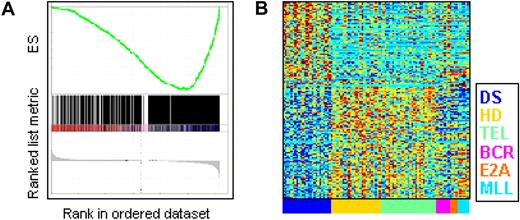
We first explored the extent of similarity between DS-ALLs and other defined BCP-ALL genetic subtypes using in the analysis the 1500 probe sets with the highest standard deviation among the AIEOP samples. Two unsupervised analysis algorithms were used: Sorting Points Into Neighborhoods (SPIN),16 which places samples with similar expression profiles near each other (Figure 1A), and Principal Component Analysis (PCA; MATLAB 7.4 software [Mathworks]; Figure 1B). Both gave similar results.
Unsupervised analysis of the AIEOP dataset. (A) Samples' Euclidian distance matrix. The color in each entry (y,x), where x,y = 1, …, 97 (the number of samples), represents the Euclidian distance between the expression profiles of samples x and y. It was measured after centering and normalization of each sample's expression, using 1500 probe sets with highest standard deviation. The samples are ordered by SPIN along both the x-axis and y-axis. The color bars next to both axes represent the different ALL subtypes, listed on the right of the figure. The blue marks at bottom specify DS-ALL samples with mutant JAK2 (J2m), and the red marks specify samples with high CRLF2 expression levels (CRLF2) (see “Aberrant expression of the cytokine receptor CRLF2 in DS-ALLs” section on CRLF2). (B) Projection of all samples onto the first 3 principle components of the expression. DS indicates Down syndrome ALL; J2m, Down syndrome ALL with mutated JAK2 R683; HD, high hyperdiploid; TEL, TEL-AML1; BCR, BCR-ABL; E2A, E2A-PBX1; and MLL, MLL-AF4.
Unsupervised analysis of the AIEOP dataset. (A) Samples' Euclidian distance matrix. The color in each entry (y,x), where x,y = 1, …, 97 (the number of samples), represents the Euclidian distance between the expression profiles of samples x and y. It was measured after centering and normalization of each sample's expression, using 1500 probe sets with highest standard deviation. The samples are ordered by SPIN along both the x-axis and y-axis. The color bars next to both axes represent the different ALL subtypes, listed on the right of the figure. The blue marks at bottom specify DS-ALL samples with mutant JAK2 (J2m), and the red marks specify samples with high CRLF2 expression levels (CRLF2) (see “Aberrant expression of the cytokine receptor CRLF2 in DS-ALLs” section on CRLF2). (B) Projection of all samples onto the first 3 principle components of the expression. DS indicates Down syndrome ALL; J2m, Down syndrome ALL with mutated JAK2 R683; HD, high hyperdiploid; TEL, TEL-AML1; BCR, BCR-ABL; E2A, E2A-PBX1; and MLL, MLL-AF4.
In agreement with previous studies,10 unsupervised analysis of gene expression tends to group the pediatric ALL samples according to their genetic subtypes. As can be seen in the Euclidian distance matrix in Figure 1A, the ALL subgroups that are the most homogenous (exhibiting high similarity of samples of the same subtype) are E2A-PBX1–positive ALL and MLL-AF4–positive ALL, followed by TEL-AML1–positive ALL. HD-ALL samples are also grouped together, but are relatively more distant from each other than the E2A-PBX1, MLL-AF4, and TEL-AML1 samples. Although BCR-ABL ALLs are clustered together, they are less homogeneous, consistent with previous reports.10
In contrast, DS-ALLs are very heterogeneous (Figure 1A-B). Approximately half are grouped together, relatively close to both BCR-ABL and HD-ALL. However even here, individual DS samples are further separated from each other than a typical pair of samples within the other ALL subtypes (Figure 1A-B). The other half are grouped with other ALL subtypes: 6 with TEL-AML1, 6 with HD, 2 with BCR-ABL, and 1 with E2A-PBX1. Of these 15 DS-ALLs, only 3 carried the chromosomal translocation of the subtype of ALL to which they are most similar (1 E2A-PBX1, 2 TEL-AML1) and only 1 DS-ALL sample was found to be also HD. Even the 5 DS-ALL samples with somatic mutations in JAK2 (blue boxes below Figure 1A and black circles at Figure 1B) are not clustered together.
This unsupervised gene expression analysis reveals that DS-ALLs are markedly less homogenous than the other ALL genetic subtypes. It suggests that DS is a predisposing condition to several genetic subtypes of B-cell precursor ALLs, and that unlike the myeloid leukemia of DS should not be considered as a unique molecular entity.
Genomic analysis of DS-ALL
We performed 100-K single nucleotide polymorphism array analysis of 34 paired diagnosis and remission samples (15 DS-, 9 HD-, and 10 TEL-AML ALLs, supplemental Methods). Copy number and loss-of-heterozygosity analyses (supplemental Figure 3; supplemental Table 10) generally confirm previous reports5,17 that deletions are more common in DS and TEL-AML1 compared with HD-ALLs. The frequency of deletions in genes regulating normal B-lymphoid development (supplemental Table 9) in DS-ALL was 53%, slightly higher than the 40% reported for of BCP-ALL.18 Recently, deletions in the IKZF1 gene were reported in the majority of patients with BCR/ABL and “BCR/ABL-like” ALL.19,20 Because most of these deletions involve only a subset of exons (most commonly exons 4-7), the 100-K single nucleotide polymorphism platform is inadequate to detect these abnormalities. Therefore, 38 additional diagnostic DS-ALL specimens were screened for IKZF1 deletions by PCR analysis as previously reported.21 Monoallelic IKZF1 deletions were identified in 9 patients (24%; supplemental Table 8 and supplemental Figure 3). Thus the frequency of deletions in B-cell differentiation genes, including IKZF1, in DS-ALL is similar to other non–BCR-ABL subtypes of BCP-ALL.
DS-ALL gene expression profile
We hypothesized that, despite their heterogeneity, DS-ALLs share a common gene expression signature. We reasoned that by comparing the gene expression in DS-ALL to the relatively similar groups, HD and TEL-AML1, we could potentially isolate the “DS-ALL” characteristics from the other ALL characteristics that might be similar between these groups. In addition, the analysis was done in a way that only genes that differentiate DS from TEL-AML1 and from HD-ALLs are depicted. The fact that TEL-AML1 and HD-ALLs have dissimilar expression profiles (denoted by dark red entries in Figure 1A) helps to identify genes that characterize DS-ALL, and not one of the groups to which it is compared.
We first identified probe sets that had significant differential expression in DS-ALL samples, compared with both HD and TEL-AML1 ALLs in the AIEOP dataset (supplemental Methods). This “preliminary DS-ALL profile” consisted of 792 genes up-regulated and 535 genes down-regulated in DS-ALL. To check consistency with each of the other 3 gene expression datasets (“BFM,” “ICH,” and “IL”), we performed GSEA22 separately on each of the 3. A representative enrichment analysis is shown in Figure 2A; here the genes are ordered according to their DS-ALL differential expression in the ICH dataset (used as the list L; “Bioinformatics”), and the 535 genes down-regulated in DS-ALL are used as the set S, tested for enrichment. Six such analyses showed significant enrichment of the preliminary gene lists (both up- and down-regulated) obtained from AIEOP, in the other 3 datasets (Table 2). We refined our AIEOP-based lists by including only genes that showed consistent expression patterns in at least 2 of the 3 other datasets. The “refined DS-ALL profile genes” (Figure 2B) consists of 152 up-regulated and 199 down-regulated genes (supplemental Tables 4-5).
DS-ALL gene expression profile. (A) GSEA analysis on ICH dataset. Genes are ranked (bottom of panel, gray) according to their expression in DS-ALL samples versus the rest of the samples, by GSEA, using the default parameters. The members of a gene set S (here, the set of 535 genes down-regulated in DS-ALL, AIEOP data) are tested: are they randomly distributed in the ranked gene list, or primarily found at the top or bottom? Occurrences of members of the gene set S in the ranked gene list are shown as vertical black lines above the ranked signature. The green curve and upper y-axis represent the enrichment score (ES) as a function of the number of ranked genes tested for enrichment of gene set S. See supplemental Methods for full details. (B) Expression levels of the genes from the refined DS-ALL lists, measured on the AIEOP dataset. Four hundred twenty-three probe sets that belong to the refined DS-ALL profile gene lists are centered and normalized. Values for each individual case are represented by a color, with red representing deviation above the mean and blue representing deviation below the mean. The colors along the x-axis represent the different ALL subtypes, listed on the right of the plot. DS indicates Down syndrome ALL; HD, high hyperdiploid; TEL, TEL-AML1; BCR, BCR-ABL; E2A, E2A-PBX1; and MLL, MLL-AF4.
DS-ALL gene expression profile. (A) GSEA analysis on ICH dataset. Genes are ranked (bottom of panel, gray) according to their expression in DS-ALL samples versus the rest of the samples, by GSEA, using the default parameters. The members of a gene set S (here, the set of 535 genes down-regulated in DS-ALL, AIEOP data) are tested: are they randomly distributed in the ranked gene list, or primarily found at the top or bottom? Occurrences of members of the gene set S in the ranked gene list are shown as vertical black lines above the ranked signature. The green curve and upper y-axis represent the enrichment score (ES) as a function of the number of ranked genes tested for enrichment of gene set S. See supplemental Methods for full details. (B) Expression levels of the genes from the refined DS-ALL lists, measured on the AIEOP dataset. Four hundred twenty-three probe sets that belong to the refined DS-ALL profile gene lists are centered and normalized. Values for each individual case are represented by a color, with red representing deviation above the mean and blue representing deviation below the mean. The colors along the x-axis represent the different ALL subtypes, listed on the right of the plot. DS indicates Down syndrome ALL; HD, high hyperdiploid; TEL, TEL-AML1; BCR, BCR-ABL; E2A, E2A-PBX1; and MLL, MLL-AF4.
Pathway analysis and BCL6 signature
To identify molecular pathways that showed differential expression in the refined DS-ALL profile, we interrogated the DAVID database23 of Gene Ontology functional categories.24 Constituent genes of 8 pathways were significantly (false discovery rate [FDR] < 10%) overrepresented in the DS-ALL expression profile (Table 3 and supplemental Table 6). The most enriched pathway (P < .001) is “Response to DNA damage stimulus”: 10 of the 341 genes assigned by DAVID to this pathway are down-regulated and 6 are up-regulated in DS-ALL. One of the up-regulated genes is BCL6, with a mean fold change of 1.46 in DS-ALL compared with non DS-ALL (supplemental Table 4). BCL6 is a transcription factor expressed primarily in mature B cells at the germinal centers, where it facilitates immunoglobulin (Ig) affinity maturation by repressing the DNA damage response. It is also a known oncogene in diffuse large B-cell lymphomas.25,26
To search for evidence for BCL6 activity in the DS-ALL gene expression profile, we used the Oncomine (http://www.oncomine.org)27,28 database, in which cancer gene expression signatures derived from different expression analyses are stored as molecular concept maps. These are lists of differentially expressed genes between 2 logical groupings of normal or malignant human tissue or cell lines. We tested BCL6 direct targets and each of the 24 Oncomine molecular concept maps that involve BCL6 (supplemental Table 7) for enrichment in DS-ALL up- and down-regulated genes, and 8 of these 25 gene groups passed at false discovery rate (FDR)29 of 15% (Table 4). These include the target genes of BCL6,32 genes modified by ectopic expression of BCL6 in lymphoblastoid B cells,30 and the gene expression signature of B-cell lymphomas with oncogenic activation of BCL6.31 Hence the targets and pathways downstream to BCL6 in lymphomas and mature B cells are modified in the DS-ALL expression profile.
Aberrant expression of the cytokine receptor CRLF2 in DS-ALLs
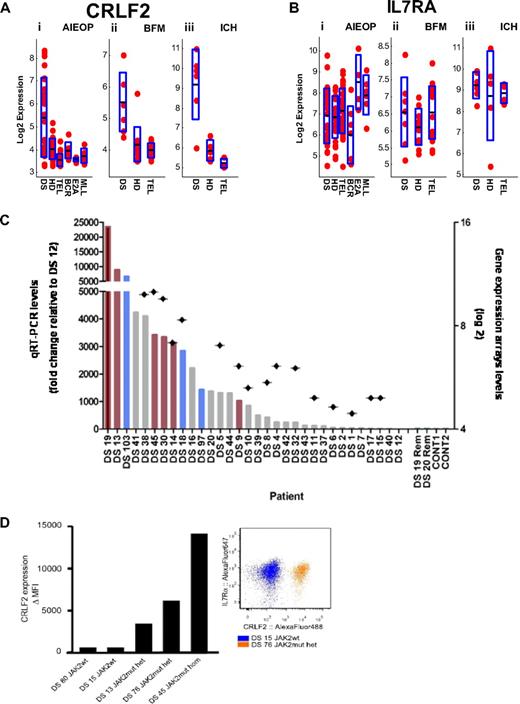
We have previously hypothesized that a cytokine receptor may be aberrantly expressed in DS-ALL and cooperate with JAK2 carrying the “lymphoid” mutation in R683.4 Examination of the DS-ALL expression signature (supplemental Table 4) reveals that the third most differentially expressed gene is CRLF2 (cytokine receptor-like factor 2, TSLPR) located at the pseudoautosomal region of the sex chromosomes. As depicted in Figure 3A, increased expression of CRLF2 was noted in 23 (62.1%) DS-ALLs of 37 samples that were hybridized to U133 family of arrays (CRLF2 is not represented on the exon arrays used in the IL dataset), compared with other ALL subtypes (see supplemental Methods). CRLF2 expression along DS-ALL versus all other ALL subtypes yielded t test P values less than .001 for AIEOP, BFM, and ICH datasets.
CRLF2 expression in DS-ALL. (A) CRLF2 expression in the AIEOP (i), BFM (ii), and ICH (iii) datasets. The y-axis represents CRLF2 log basis 2 expression. The x-axis represents the different ALL subtypes. Each point corresponds to a sample. The black line in each ALL subtype is the CRLF2 mean (log basis 2) expression in this subtype. The height of the blue rectangle in each ALL subtype is the measured standard deviation of CRLF2 (log basis 2) expression. DS-ALL versus all other ALL yielded t test P values of less than .001 for AIEOP (i), BFM (ii), and ICH (iii). DS indicates Down syndrome ALL; HD, high hyperdiploid; TEL, TEL-AML1; BCR, BCR-ABL; E2A, E2A-PBX1; and MLL, MLL-AF4. (B) IL7RA expression in the AIEOP (i), BFM (ii), and ICH (iii) datasets. There are no statistically significant differences between DS-ALLs and non–DS-ALLs. (C) Verification of CRLF2 expression levels by qRT-PCR. Bars represent qRT-PCR CRLF2 expression levels (left, y-axis: fold change relative to patient DS-12, the lowest CRLF2 expressor). Rhombuses represent gene expression array CRLF2 expression levels (right, y-axis: log basis 2). Red bars represent patients with JAK2 R683 mutation; blue bars, patients with CRLF2 F232C mutation (Figure 6). Rem indicates CRLF2 levels of available remission samples (patients DS-19 and DS-20); CONT, control CRLF2 expression levels in peripheral white blood cells of healthy donors. (D) CRLF2 and IL7RA protein expression on the surface of DS-ALL leukemic blasts. (Left panel) Delta mean fluorescence intensity of the signal detected by flow cytometry using specific anti-CRLF2 antibodies compared with background unspecific staining (“Flow cytometric analysis”), indicating an apparent association between the JAK2 mutational status and the level of expression of CRLF2 on DS-ALL blasts. (Right panel) Dot plot of 2 representative CRLF2 and IL7RA costainings. IL7RA is highly expressed on leukemic blasts independent of JAK2 mutational status and level of CRLF2 expression in all cases examined. wt indicates wild-type; mut, mutant; het, heterozygous; and hom, homozygous.
CRLF2 expression in DS-ALL. (A) CRLF2 expression in the AIEOP (i), BFM (ii), and ICH (iii) datasets. The y-axis represents CRLF2 log basis 2 expression. The x-axis represents the different ALL subtypes. Each point corresponds to a sample. The black line in each ALL subtype is the CRLF2 mean (log basis 2) expression in this subtype. The height of the blue rectangle in each ALL subtype is the measured standard deviation of CRLF2 (log basis 2) expression. DS-ALL versus all other ALL yielded t test P values of less than .001 for AIEOP (i), BFM (ii), and ICH (iii). DS indicates Down syndrome ALL; HD, high hyperdiploid; TEL, TEL-AML1; BCR, BCR-ABL; E2A, E2A-PBX1; and MLL, MLL-AF4. (B) IL7RA expression in the AIEOP (i), BFM (ii), and ICH (iii) datasets. There are no statistically significant differences between DS-ALLs and non–DS-ALLs. (C) Verification of CRLF2 expression levels by qRT-PCR. Bars represent qRT-PCR CRLF2 expression levels (left, y-axis: fold change relative to patient DS-12, the lowest CRLF2 expressor). Rhombuses represent gene expression array CRLF2 expression levels (right, y-axis: log basis 2). Red bars represent patients with JAK2 R683 mutation; blue bars, patients with CRLF2 F232C mutation (Figure 6). Rem indicates CRLF2 levels of available remission samples (patients DS-19 and DS-20); CONT, control CRLF2 expression levels in peripheral white blood cells of healthy donors. (D) CRLF2 and IL7RA protein expression on the surface of DS-ALL leukemic blasts. (Left panel) Delta mean fluorescence intensity of the signal detected by flow cytometry using specific anti-CRLF2 antibodies compared with background unspecific staining (“Flow cytometric analysis”), indicating an apparent association between the JAK2 mutational status and the level of expression of CRLF2 on DS-ALL blasts. (Right panel) Dot plot of 2 representative CRLF2 and IL7RA costainings. IL7RA is highly expressed on leukemic blasts independent of JAK2 mutational status and level of CRLF2 expression in all cases examined. wt indicates wild-type; mut, mutant; het, heterozygous; and hom, homozygous.
CRLF2 is known to dimerize with IL7RA to form the heterodimeric receptor for thymic stromal lymphopoietin (TSLP).33 Whereas CRLF2 is aberrantly expressed in DS-ALLs (Figure 3A), expression of IL7RA is similar in the different ALL subtypes (Figure 3B).
To validate the findings of the expression arrays and to analyze additional DS-ALL samples, we measured the expression of CRLF2 by qRT-PCR in 32 patients (Figure 3C). Microarray data were available for 16 of these cases. The qRT-PCR confirms the CRLF2 expression levels seen in the arrays (Pearson correlation = 0.85, P < .001). In 2 patients' CRLF2 expression was analyzed in RNA derived from diagnostic and remission bone marrows and was seen only in the diagnostic sample. In one patient, similar CRLF2 expression levels were seen in bone marrow samples from diagnosis and relapse (supplemental Figure 2). Altogether, 33 (62.3%) of 53 DS-ALL patients analyzed by either qRT-PCR or microarrays overexpressed CRLF2. The surface expression of the CRLF2 protein was also verified on 4 samples by flow cytometry (Figure 3D). IL7RA is also expressed on the leukemic blasts independent of CRLF2 expression.
Recently, Russell et al13 reported aberrant expression of CRLF2 caused by either chromosomal translocations to the IGH@ locus or interstitial deletions upstream to CRLF2 juxtaposing CRLF2 with the P2RY8 regulatory elements in approximately 5% of childhood ALLs. To examine whether the increased CRLF2 expression in our specimens was caused by the same genomic aberrations, 12 available diagnostic DS-ALL samples overexpressing CRLF2 were analyzed by FISH (Figure 4A). IGH@ translocations were seen in 4 specimens and interstitial deletions in 7. In the remaining sample (no. DS-32; supplemental Table 2), in which the CRLF2 expression level was just above the threshold, the FISH pattern of CRLF2 appeared normal. Further evidence supporting the presence of the deletions is provided by a statistically significant inverse correlation between CRLF2 and P2RY8 expression (P = .02, Figure 4B).
Genomic analysis of CRLF2 aberrations. (A) FISH analysis of DS-ALL expressing CRLF2 (i-ii) IGH@ CRLF2 translocation, patient DS-85: (i) Metaphase showing a positive result with the LSI IGH@ break-apart rearrangement probe (Abbott Molecular): normal chromosome 14 (yellow arrow) derived chromosome 14 (red arrow), derived X chromosome (green arrow). (ii) Interphase nucleus from the same patient hybridized with the homegrown CRLF2 probe showing a split signal pattern, 1R1G1F, confirming its involvement in the translocation (1 fusion signal [yellow arrow], 1 red signal [red arrow], and 1 green signal [green arrow]). (iii-iv) CRLF2 microdeletion, patient DS-82. (iii) Interphase nucleus hybridized with the IGH@ probe showing the normal 0R0G2F signal pattern confirming the presence of 2 normal copies of IGH@. (iv) Interphase nucleus from the same patient hybridized with the homegrown CRLF2 probe showing the deletion of the green portion of the probe (1 red signal [red arrow] and 1 fusion signal [yellow arrow]) denoting the presence of a centromeric interstitial deletion. (B) CRLF2 and P2RY8 expression in DS-ALL samples. Centered and normalized log basis 2 expression of CRLF2 and P2RY8 along DS-ALL samples in AIEOP dataset. Values for each individual case are represented by a color, with red representing deviation above the mean and blue representing deviation below the mean. The samples are sorted using SPIN. Pearson correlation between CRLF2 and P2RY8: −0.45 (P = .02). (C) Detection of the P2RY8-CRLF2 fusion transcript. (i) Schematic representation of the deletion breakpoint region at the telomeric end of chromosome X/Y with gene locations. The dashed lines represent the genomic deletion leading to the fusion of the first noncoding exon of P2RY8 and to the first (coding) exon of CRLF2. (iii) RT-PCR experiments on cDNA of DS patients. (Lanes 1-3) DS diagnostic ALL samples (DS93 and DS82 with FISH determined deletion and DS92 with FISH determined IgH@ translocation). (Lane 4) Blank. The 3 patient samples were positive for ABL amplification (not shown). Primer sets used are (A) P2RY8 F01/CRLF2 R01; (B) P2RY8 F01/ CRLF2 R02; (C) P2RY8 F01/CRLF2 R03 shown on the sequence on the left (ii). Chimeric transcripts are present in the first 2 lanes of each set. (ii) Nucleotide sequencing of the largest PCR fragment confirming the fusion transcript; vertical lines indicate exon boundaries; the arrowhead indicates the P2RY8/CRLF2 transcript junction. As seen, the fusion is just upstream to the ATG of CRLF2. Reference sequences are P2RY8-001 (ENST00000381297) and CRLF2-001 (ENST00000400841). Boxed sequence around the transcript junction is represented in the electropherogram on the right lower side (iv).
Genomic analysis of CRLF2 aberrations. (A) FISH analysis of DS-ALL expressing CRLF2 (i-ii) IGH@ CRLF2 translocation, patient DS-85: (i) Metaphase showing a positive result with the LSI IGH@ break-apart rearrangement probe (Abbott Molecular): normal chromosome 14 (yellow arrow) derived chromosome 14 (red arrow), derived X chromosome (green arrow). (ii) Interphase nucleus from the same patient hybridized with the homegrown CRLF2 probe showing a split signal pattern, 1R1G1F, confirming its involvement in the translocation (1 fusion signal [yellow arrow], 1 red signal [red arrow], and 1 green signal [green arrow]). (iii-iv) CRLF2 microdeletion, patient DS-82. (iii) Interphase nucleus hybridized with the IGH@ probe showing the normal 0R0G2F signal pattern confirming the presence of 2 normal copies of IGH@. (iv) Interphase nucleus from the same patient hybridized with the homegrown CRLF2 probe showing the deletion of the green portion of the probe (1 red signal [red arrow] and 1 fusion signal [yellow arrow]) denoting the presence of a centromeric interstitial deletion. (B) CRLF2 and P2RY8 expression in DS-ALL samples. Centered and normalized log basis 2 expression of CRLF2 and P2RY8 along DS-ALL samples in AIEOP dataset. Values for each individual case are represented by a color, with red representing deviation above the mean and blue representing deviation below the mean. The samples are sorted using SPIN. Pearson correlation between CRLF2 and P2RY8: −0.45 (P = .02). (C) Detection of the P2RY8-CRLF2 fusion transcript. (i) Schematic representation of the deletion breakpoint region at the telomeric end of chromosome X/Y with gene locations. The dashed lines represent the genomic deletion leading to the fusion of the first noncoding exon of P2RY8 and to the first (coding) exon of CRLF2. (iii) RT-PCR experiments on cDNA of DS patients. (Lanes 1-3) DS diagnostic ALL samples (DS93 and DS82 with FISH determined deletion and DS92 with FISH determined IgH@ translocation). (Lane 4) Blank. The 3 patient samples were positive for ABL amplification (not shown). Primer sets used are (A) P2RY8 F01/CRLF2 R01; (B) P2RY8 F01/ CRLF2 R02; (C) P2RY8 F01/CRLF2 R03 shown on the sequence on the left (ii). Chimeric transcripts are present in the first 2 lanes of each set. (ii) Nucleotide sequencing of the largest PCR fragment confirming the fusion transcript; vertical lines indicate exon boundaries; the arrowhead indicates the P2RY8/CRLF2 transcript junction. As seen, the fusion is just upstream to the ATG of CRLF2. Reference sequences are P2RY8-001 (ENST00000381297) and CRLF2-001 (ENST00000400841). Boxed sequence around the transcript junction is represented in the electropherogram on the right lower side (iv).
To test whether the deletion caused a fusion between the P2RY8 and CRLF2, we performed RT-PCR with primers derived from both genes (supplemental Table 3). A transcript fusing the first noncoding exon of P2RY8 and the first exon of CRLF2 prior to the ATG was detected in the 2 DS patients with the deletion detected by FISH but not in the patient with the IgH@ translocation (Figure 4C). A similar chimeric transcript was described in a single patient with splenic lymphoma, fusing P2RY8 to SOX5 resulting in overexpression of SOX5.34 We extended the analysis and identified the chimeric transcript in 7 of 10 patients with overexpression of CRLF2 and in none of 8 samples with no expression of CRLF2 (supplemental Table 2). Thus, consistent with the FISH findings,13 the interstitial deletion is more common than the IgH@ translocation.
To explore the effect of CRLF2 on gene expression, we compared the 30% of DS-ALLs with the highest CRLF2 expression and the 30% of DS-ALLs with the lowest CRLF2 expression in the AIEOP database. Only 5 probe sets passed FDR of 30%, with CRLF2 being 1 of the 5 (Table 5). This is consistent with the finding that samples that overexpress CRLF2 (red marks in Figure 1A) do not cluster separately from DS-ALLs that do not express CRLF2. Interestingly, the IGJ gene that differentiates these 2 groups (fold change, 36.4; Table 5) is also the most differentiating gene between DS-ALL and non–DS-ALL in our datasets (supplemental Table 4).
Clinical significance of CRLF2 expression in DS-ALL
Clinically, children with high/medium expression of CRLF2 were diagnosed younger (Table 6) than children with no/low expression of CRLF2 (5.56 vs 9.87 years, P = .004).
No significant differences between the 2 groups regarding their sex or white blood cell count at diagnosis were found. Patients expressing CRLF2 tended to have a lower probability for event-free survival (supplemental Figure 4, P = .12 log-rank test).
Cooperation between JAK2 R683 mutations and CRLF2 aberrant expression
Among the 53 DS-ALL samples for which CRLF2 expression was available, 10 had somatic mutations in JAK2 R683. We identified chimeric P2RY8-CRLF2 transcripts in 3 additional patients with JAK2 R683 mutations (supplemental Table 2). Thus all mutations occurred in specimens with aberrant expression of CRLF2, supporting our initial hypothesis that CRLF2 may act as type I cytokine receptor for mutated JAK2.
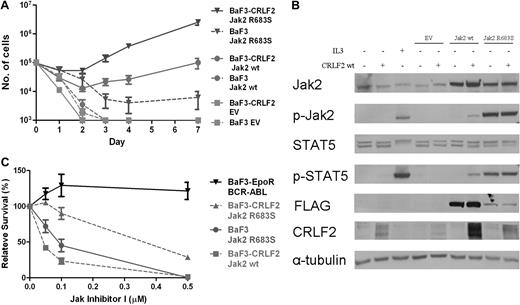
To examine whether CRLF2 and mutated JAK2 cooperate, we generated BaF3 cells that express hCRLF2 (BaF3-CRLF2) and transduced both BaF3 and BaF3-CRLF2 cells with wild-type mJak2-FLAG, R683S mJak2-FLAG, and empty vector. As depicted in Figure 5A, there was synergism between CRLF2 and both wt Jak2 and R683S mutated Jak2, with the best cytokine-independent growth observed in cells expressing CRLF2 and the mutated Jak2. These functional effects on cell growth are reflected in protein analysis of the JAK/STAT pathway (Figure 5B). Interestingly, despite identical levels of CRLF2 at the time of transduction, the levels of CRLF2 were consistently higher in cells transduced with Jak2 compared with empty vector or parental cells. Examination of Stat5 and Jak2 phosphorylation 5 hours after cytokine withdrawal reveals that when CRLF2 was expressed, phosphorylation levels in cells transduced with wt Jak2 were increased, whereas no change was observed in the already high phosphorylation levels in cells expressing the mutated Jak2. The marked advantage in cytokine-independent growth rate of cells coexpressing CRLF2 and R683S Jak2 despite similar Stat5 phosphorylation may indicate the involvement of additional signaling pathways.
Functional significance of CRLF2 expression. (A) Cytokine withdrawal assay of BaF3 and BaF3-CRLF2 cells infected with either empty vector (EV), mouse FLAG-Jak2 wild-type (wt), or mouse FLAG-Jak2 R683S. Error bars represent SE. (B) Constitutive activation of the Jak/Stat5 pathway in BaF3 and BaF3-CRLF2 cells expressing mouse FLAG-Jak2 wild-type (wt) or R683S, after 5 hours of cytokine deprivation. IL3+ indicates cells harvested after 5 hours of interleukin-3 deprivation followed by 15 minutes of interleukin-3 stimulation. (C) Effect of JAK inhibitor I on growth of BaF3 cells expressing Jak2 R683S and BaF3-CRLF2 cells expressing either wt Jak2 or Jak2 R683S.
Functional significance of CRLF2 expression. (A) Cytokine withdrawal assay of BaF3 and BaF3-CRLF2 cells infected with either empty vector (EV), mouse FLAG-Jak2 wild-type (wt), or mouse FLAG-Jak2 R683S. Error bars represent SE. (B) Constitutive activation of the Jak/Stat5 pathway in BaF3 and BaF3-CRLF2 cells expressing mouse FLAG-Jak2 wild-type (wt) or R683S, after 5 hours of cytokine deprivation. IL3+ indicates cells harvested after 5 hours of interleukin-3 deprivation followed by 15 minutes of interleukin-3 stimulation. (C) Effect of JAK inhibitor I on growth of BaF3 cells expressing Jak2 R683S and BaF3-CRLF2 cells expressing either wt Jak2 or Jak2 R683S.
To test whether the cells expressing CRLF2 and/or either wt or R683 mutated Jak2 depend on activated JAK signaling, we incubated BaF3 cells transduced with the different vectors cultured without IL3 in the presence of different concentrations of JAK inhibitor l (Figure 5C). Although BaF3 cells transduced with CRLF2/Jak were more sensitive to the inhibitor compared with the control cells expressing BCR-ABL (P = .04, analysis of variance), the cells expressing CRLF2 and mutated Jak2 were the least sensitive.
Activating mutations of CRLF2 in DS-ALL
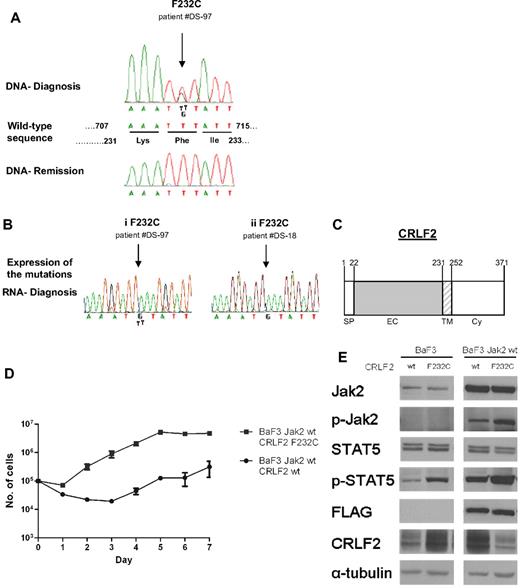
To identify additional events leading to CRLF2 activation, we screened 87 diagnostic DS-ALL samples for mutations in CRLF2 (Figure 6). In addition to polymorphisms V136M and V244M that were present also in remission samples and in healthy controls, we identified in 3 patients a somatic mutation replacing phenylalanine 232, located at the juxtamembranous domain, with cysteine (F232C). Genomic data were available for one of the patients (DS-97) who displayed the P2YR8-CRLF2 transcript. Although F232C induced constitutive Stat5 phosphorylation in cytokine-deprived BaF3 cells (Figure 6E), it did not provide a consistent survival advantage. Whereas during the first few days after cytokine withdrawal more cells expressing F232C CRLF2 were alive compared with cells expressing wt CRLF2, at day 7 almost all BaF3 cells were dead (not shown). To examine the collaboration with wt Jak2, BaF3 cells stably expressing wt Jak2 were transduced with retroviral vectors expressing either wt CRLF or F232C CRLF2 (Figure 6D-E). In the presence of exogenous wt Jak2, there was an approximately 15-fold increase in the growth rate of cells expressing the mutant CRLF2 compared with those expressing wt CRLF2 (P = .02, paired t test). Together these observations demonstrate that the F232C CRLF2 activates Jak/Stat signaling and cooperates with Jak2 to provide significant growth advantage in a cytokine-deprived environment.
Mutations of CRLF2 in patients with Down syndrome–associated acute lymphoblastic leukemia. (A) Example of sequences depicting the F232C in CRLF2. The F232C (arrowed) is present at diagnosis but not in remission. The wild-type sequence denotes positions of both nucleotides and amino acids. (B) Expression of CRLF2 F232C mutation. Examples of 2 patients: in one (i) both alleles, wild-type and mutated, are expressed, whereas in the other (ii) only the mutated allele is expressed. (C) Schematic presentation of CRLF2. SP indicates signal peptide; EC, extracellular region; TM, transmembrane region; and Cy, cytoplasmic region. Numbers indicate amino acid position. (D) Cytokine withdrawal assay of BaF3 cells stably expressing wild-type mouse FLAG-Jak2 that were transduced with either wild-type human CRLF2 or human CRLF2 F232C. Error bars represent SE. (E) Constitutive activation of the Jak/Stat5 pathway in BaF3 cells expressing wild-type mouse FLAG-Jak2 and either wild-type human CRLF2 (wt) or human CRLF2 F232C (F232C), after 5 hours of cytokine deprivation.
Mutations of CRLF2 in patients with Down syndrome–associated acute lymphoblastic leukemia. (A) Example of sequences depicting the F232C in CRLF2. The F232C (arrowed) is present at diagnosis but not in remission. The wild-type sequence denotes positions of both nucleotides and amino acids. (B) Expression of CRLF2 F232C mutation. Examples of 2 patients: in one (i) both alleles, wild-type and mutated, are expressed, whereas in the other (ii) only the mutated allele is expressed. (C) Schematic presentation of CRLF2. SP indicates signal peptide; EC, extracellular region; TM, transmembrane region; and Cy, cytoplasmic region. Numbers indicate amino acid position. (D) Cytokine withdrawal assay of BaF3 cells stably expressing wild-type mouse FLAG-Jak2 that were transduced with either wild-type human CRLF2 or human CRLF2 F232C. Error bars represent SE. (E) Constitutive activation of the Jak/Stat5 pathway in BaF3 cells expressing wild-type mouse FLAG-Jak2 and either wild-type human CRLF2 (wt) or human CRLF2 F232C (F232C), after 5 hours of cytokine deprivation.
Discussion
Here we report the results of a genome-wide study of DS-ALL based on a dataset of unprecedented size. Unexpectedly, the molecular phenotype obtained by gene expression profiling is strikingly less homogeneous in DS-ALL than any of the common genetic subtypes of childhood BCP-ALLs. However, despite this heterogeneity, we describe a major feature that is shared by up to two-thirds of the patients—the aberrant expression of the wt or mutated cytokine receptor CRLF2 and its association with mutations in JAK2.
That DS-ALL is less uniform than the specific DS-associated myeloid leukemia has been suggested by a large cytogenetic study performed by the International BFM Study Group.2 However, neither that study nor the genomic analysis reported here or previously5,17 explains the level of inhomogeneity in gene expression. Even those DS-ALLs that clustered together were not similar to each other. Such heterogeneity suggests that unlike the common aberrations of childhood ALL (TEL-AML1, hyperdiploidy, E2A-PBX1, etc), constitutional trisomy 21 is not a typical initiating event. Rather, DS is a predisposing condition to multiple genetic subtypes of BCP-ALLs.
To identify genes and pathways common to DS-ALLs, we have generated a DS-ALL gene expression signature, exploiting the advantage of having several datasets. CRLF2 is 1 of the 3 genes that most differentiates between DS-ALLs and non–DS-ALLs. Confirming the expression of CRLF2 RNA and protein in DS-ALLs and extending these observations to patients for whom array data were not available, we observed increased expression of CRLF2 in 62% of 53 patients with DS-ALL. These data are corroborated by the recent report describing IGH@ translocations or interstitial deletions upstream of CRLF2 in 5% of nonselected childhood ALL, and in 35 (52%) of 68 DS-ALLs consecutively enrolled in United Kingdom treatment protocols.13
We report that the interstitial deletion results in fusion transcript in which the first noncoding exon of P2YR8 fuses to the coding region of CRLF2, thereby driving CRLF2 expression by the P2YR8 promoter. A similar mechanism was reported in a single patient with splenic lymphoma and P2YR8-SOX5 fusion34 and is reminiscent of the common SIL-SCL (STIL-TAL1) rearrangement in T-ALL.35 Cloning of the genomic breakpoints is required to determine whether, like SIL-SCL, the deletion is caused by aberrant V(D)J activity.
Although we do not have genomic data for all CRLF2-expressing samples, the FISH and RT-PCR results of 17 of 33 specimens overexpressing CRLF2, the inverse correlation between CRLF2 and P2YRY8 expression, and the similar frequency of CRLF2 overexpression in our and Russell et al's13 2 independent cohorts suggest that most, if not all, aberrant CRLF2 expression is caused by genomic rearrangements.
CRLF2 dimerizes with IL7RA to form the receptor to thymic stromal-derived lymphopoietin (TSLP), an epithelial-derived cytokine that plays a role in inflammation and lymphoid development.14,36-38 The expression of IL7RA on the leukemic blasts suggests that some of the aberrantly expressed CRLF2 may interact with IL7RA and form a TSLP receptor on the leukemic cells. However, we also demonstrate that CRLF2 cooperates with Jak2 to transform BaF3 cells lacking expression of IL7RA (Figure 5A and supplemental Figure 5). This suggests that CRLF2 may act independently of IL7RA, possibly through homodimerization similar to other type I cytokine receptors.
We report an unusual cooperation between CRLF2 and ectopically expressed wt Jak2 in BaF3 cells, a phenomenon not observed with other type I cytokine receptors such as erythropoietin receptor (EPOR) or thrombopoietin receptor (TPOR). CRLF2 is an atypical type I cytokine receptor that contains only one of the 2 “boxes” that mediate binding of JAK enzymes and only one tyrosine in its C-terminal domain. Hence it is a weak activator of JAK2.39 This may explain the requirements for higher levels of Jak2 for activation of the Jak/Stat pathway. Interestingly, the levels of CRLF2 were higher in the presence of ectopically expressed wt or mutated Jak2. Positive regulation of the expression of a type I cytokine receptor by JAK2 and Tyk2 was previously reported.40-42 Thus one mechanism by which Jak2 may cooperate with CRLF2 is by increasing the expression of the latter.
We observed 2 acquired events associated with the increased expression of CRLF2. The most common event is activating “lymphoid” somatic mutation in JAK2. All DS-ALLs specimens with JAK2 mutations in our series and in the cohort reported by Russell et al13 had aberrant expression of CRLF2, strongly implying that CRLF2 is the cytokine receptor cooperating with R683 mutated JAK2. Indeed, in BaF3 cytokine weaning assays, only the combination of CRLF2 and mutated Jak2 led to a robust cytokine-independent growth, demonstrating for the first time that these 2 proteins cooperate in providing growth and survival advantage.
The second less common event is an activating mutation in CRLF2. Yoda et al43 reported an E40G activating somatic mutation in CRLF2 in a single patient with adult BCP-ALL. We now found that 3 of the 33 patients with DS-ALL overexpressing CRLF2 have a somatic mutation replacing phenylalanine in the juxtamembrane position 232 by cysteine. This mutation caused constitutive phosphorylation of Stat5 associated with robust cytokine-independent growth of BaF3 cells ectopically transduced with wt Jak2. Introduction of cysteines in this region in the erythropoietin receptor, another type I cytokine receptor signaling through JAK2, caused its constitutive activation by enhancing ligand-independent dimerization.44
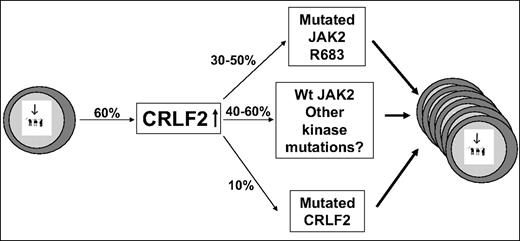
Although several scenarios may be possible, a reasonable model (Figure 7) is that the overexpression of CRLF2 is the first event occurring in approximately 60% of DS-ALL patients. The expanded preleukemic clone then acquires additional genetic aberrations, among them an activating mutation in JAK2 or CRLF2 or thus far unidentified events that may involve the JAK/STAT pathway. This model explains 3 key observations: (1) All samples with mutated JAK2 and the only evaluable patient with mutated CRLF2 also had aberrant CRLF2 expression. (2) Many CRLF2-overexpressing samples do not have mutation in JAK2. (3) In one reported patient,13 an aberrant CRLF2 genomic rearrangement was present at diagnosis whereas mutant JAK2 was present only in the relapse sample.
CRLF2 in DS-ALL: a model. Increased CRLF2 expression caused by genomic aberration is followed by progression event consisting of activating mutations in CRLF2, in JAK2, or other alterations in yet unidentified kinases. The percentages in the figure are approximations based on combination of the data in our paper and in the paper by Russell et al.13
CRLF2 in DS-ALL: a model. Increased CRLF2 expression caused by genomic aberration is followed by progression event consisting of activating mutations in CRLF2, in JAK2, or other alterations in yet unidentified kinases. The percentages in the figure are approximations based on combination of the data in our paper and in the paper by Russell et al.13
The most intriguing question is why there is a dramatic 10-fold increase in genomic lesions causing CRLF2 overexpression in DS (60% in DS-ALL compared with 5% in sporadic ALL) and how this relates to trisomy 21. Only a single Hsa21 gene, SON, was included in the DS-ALL signature and it was only slightly (1.3) up-regulated (supplemental Table 4). Indeed, we found no major difference in the gene expression from the trisomic chromosome 21 between DS-ALL and HD-ALL (data not shown). Yet our data suggest that DS-ALL and HD-ALL are to a great extent different leukemias. There are obvious fundamental differences between constitutional and acquired trisomy,45 such as the developmental stage in which the trisomy occurs and the fact that a constitutional trisomy is present both in the leukemia cells and in their microenvironment.
Regardless of the role of the constitutional trisomy, our data generate an intriguing hypothesis. We observe a significant enrichment in DNA damage and repair genes in DS-ALL and identify increased expression and clear “footprint” of BCL6 in these leukemias. BCL6 regulates the germinal center B-cell maturation, through its effects on the DNA damage response. Recent studies by Duy et al46 suggest for the first time a role for BCL6 in BCP-ALL. We speculate that DS may predispose to ALL through B-cell lymphocytic specific genomic instability involving BCL6. The signatures of BCL6 and the DNA damage response pathway may be related to previous reports on impaired cellular response to DNA damage in DS47 and to the increased prevalence of IgH@ chromosomal translocation in DS-ALL.13,48 At present, however, it is impossible to determine whether the BCL6 signature precedes or follows the CRLF2 rearrangements. As high expression of CRLF2 blocks B-cell differentiation,13 one cannot exclude the possibility that it causes a developmental arrest of the preleukemic cell in a stage in which BCL6 is active. Distinguishing between these 2 hypotheses will require the identification and study of preleukemic cells in children with DS.
Finally, our data imply that therapeutics targeting JAK/STAT signaling may be of potential benefit to the majority of DS-ALLs not limited only to those with mutated JAK2. Although we demonstrate that BaF3 cells coexpressing CRLF2 and mutated Jak2 are more susceptible to JAK inhibitor l than cells transformed with BCR-ABL, they were relatively resistant in comparison with cells transformed only with mutated Jak2. This preliminary observation requires further testing in primary leukemic cells. It may indicate that targeting other pathways activated by CRLF2 or the use of anti–CRLF2-specific antibodies will synergize with JAK2 inhibitors in treatment of DS-ALL and non–DS-ALL with aberrant CRLF2 expression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr T. Enver for supplying the pHRSINCSGW lentivirus plasmid and Dr Kitamura for providing the CRLF2 expression construct. We thank N. Amariglio, G. Basso, V. Binder, A. Biondi, S. Bresolin, O. Haas, J. Harbott, M. Grazia, G. Martinelli, M. Schrappe, S. Orkin, D. Weinstock, A. Yitzhaky, Y. Shinar, A. Ovadia, and the members of E. Domany's and S. Izraeli's groups, in particular Y. Birger and E. Ben-Meir, for their important contributions to the study.
Funding for this study was provided by the Israel Ministry of Science (E.D. and S.I.), Israeli Ministry of trade (D.B. and S.I.), Israeli Ministry of Health (S.I.), German Israeli Foundation (S.I. and A.B.), Israeli Science Foundation and Morasha program (S.I.), US Israel Binational Science Foundation (S.I.), Children with Leukemia UK (S.I.), Curtis Katz (S.I.), Wolfson Foundation (E.D. and S.I.), Foulkes Foundation (L.H. and I.G.), The Ridgefield Foundation (E.D.), Converging Technologies Program (L.H.), Constantiner Institute for Molecular Genetics (I.G.), Hans Altschüler-Stiftung (S.I. and J.P.B.), Swiss National Science Foundation (J.P.B.), Leukemia Research Fund, UK (C.H. and L.J.R.), PRIN/Programmi di ricerca di Relevante Interesse Nazionale, Rome (GteK), European LeukemiaNet, AIRC, AIL, Fondazione Del Monte di Bologna e Ravenna, FIRB 2006 (II). Reagents for the AIEOP GEP study were supplied by Roche Molecular Diagnostics in the framework of the MILE study.49
This study has been performed as partial fulfillment of the requirement for a PhD degree from the Tel Aviv University Faculty of Medicine to L.H., I.G., and C.S.
Authorship
Contribution: S.I., E.D., J.-P.B., H.K., G. Cazzaniga, G.K., G.R., and M. Stanulla conceived and planned the study; G. Cazzaniga, G. Cario, M. Stanulla, S.S., A.Y., and A.B. provided reagents and/or important data; E.V., I.G., G. Cazzaniga, M. Schmitz, J.C., R.S., I.I., C.S., S.Z., L.J.R., C.J.H., B.B., D.B., J.-P.B., G.K., and S.I. performed and/or analyzed experiments; L.H. and E.D. performed all the bioinformatics analysis; L.H., I.G., S.I., and E.D. wrote the paper with contributions by M. Schmitz, I.I., R.S., C.S., S.Z., G. Cazzaniga, S.S., and L.J.R. S.I. assumes full responsibility for the content of the paper including supplemental files.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shai Izraeli, Pediatric Hematology Oncology Dept, Sheba Medical Center, Tel Hashomer Ramat Gan 52621 Israel; e-mail: sizraeli@sheba.health.gov.il.
References
Author notes
*L.H., E.V., and I.G. are co–first authors.
†G.t.K., J.-P.B., E.D., and S.I. are co–senior authors.
Supplemental data
241 probe sets corresponding to 199 genes. Probe set Id is of U133 Plus 2.0 array in Italy dataset. Fold change: ratio between mean expression in DS ALL and mean expression in TEL-AML1 and HHD ALL (together).Additional supplemental tables and supplemental figures can be found here.

![Figure 4. Genomic analysis of CRLF2 aberrations. (A) FISH analysis of DS-ALL expressing CRLF2 (i-ii) IGH@ CRLF2 translocation, patient DS-85: (i) Metaphase showing a positive result with the LSI IGH@ break-apart rearrangement probe (Abbott Molecular): normal chromosome 14 (yellow arrow) derived chromosome 14 (red arrow), derived X chromosome (green arrow). (ii) Interphase nucleus from the same patient hybridized with the homegrown CRLF2 probe showing a split signal pattern, 1R1G1F, confirming its involvement in the translocation (1 fusion signal [yellow arrow], 1 red signal [red arrow], and 1 green signal [green arrow]). (iii-iv) CRLF2 microdeletion, patient DS-82. (iii) Interphase nucleus hybridized with the IGH@ probe showing the normal 0R0G2F signal pattern confirming the presence of 2 normal copies of IGH@. (iv) Interphase nucleus from the same patient hybridized with the homegrown CRLF2 probe showing the deletion of the green portion of the probe (1 red signal [red arrow] and 1 fusion signal [yellow arrow]) denoting the presence of a centromeric interstitial deletion. (B) CRLF2 and P2RY8 expression in DS-ALL samples. Centered and normalized log basis 2 expression of CRLF2 and P2RY8 along DS-ALL samples in AIEOP dataset. Values for each individual case are represented by a color, with red representing deviation above the mean and blue representing deviation below the mean. The samples are sorted using SPIN. Pearson correlation between CRLF2 and P2RY8: −0.45 (P = .02). (C) Detection of the P2RY8-CRLF2 fusion transcript. (i) Schematic representation of the deletion breakpoint region at the telomeric end of chromosome X/Y with gene locations. The dashed lines represent the genomic deletion leading to the fusion of the first noncoding exon of P2RY8 and to the first (coding) exon of CRLF2. (iii) RT-PCR experiments on cDNA of DS patients. (Lanes 1-3) DS diagnostic ALL samples (DS93 and DS82 with FISH determined deletion and DS92 with FISH determined IgH@ translocation). (Lane 4) Blank. The 3 patient samples were positive for ABL amplification (not shown). Primer sets used are (A) P2RY8 F01/CRLF2 R01; (B) P2RY8 F01/ CRLF2 R02; (C) P2RY8 F01/CRLF2 R03 shown on the sequence on the left (ii). Chimeric transcripts are present in the first 2 lanes of each set. (ii) Nucleotide sequencing of the largest PCR fragment confirming the fusion transcript; vertical lines indicate exon boundaries; the arrowhead indicates the P2RY8/CRLF2 transcript junction. As seen, the fusion is just upstream to the ATG of CRLF2. Reference sequences are P2RY8-001 (ENST00000381297) and CRLF2-001 (ENST00000400841). Boxed sequence around the transcript junction is represented in the electropherogram on the right lower side (iv).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/5/10.1182_blood-2009-08-235408/4/m_zh89990947790004.jpeg?Expires=1769127906&Signature=btZkEMoQIq7vBuB25D3TS~3djy-zzz-Kdojl8qPH68L4IIgtL2QQujmML2SjPqZhjVKBFYb-YHEtVDy~q0FYp1EaPKg3Dql0WPt6lY2Zg~6ES0QYCSwAJoxlYea1z3Pm3tOF16wrTSpJVbMcQbaT37fvl-wyzHMzTs38eaW3~v5Es-MpauvJUkJS-KO3ECiFnDnRpsJ8FpeP8WlDj5q9e07O0C7HW5TA9vZ~DhKdUV2Y54oXLhymYKtsBW8AVNxLSATjkLaQJCqmkYakTueGU5bo-YqfoJMik6R2I1v4XTaO75-KPMYaThOXHcPu~CNKHnz4ZMlcrb0r3HnK3ntPRA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)