Abstract
In patients with multiple myeloma (MM), risk stratification by chromosomal abnormalities may enable a more rational selection of therapeutic approaches. In the present study, we analyzed the prognostic value of 12 chromosomal abnormalities in a series of 354 MM patients treated within the HOVON-65/GMMG-HD4 trial. Because of the 2-arm design of the study, we were able to analyze the effect of a bortezomib-based treatment before and after autologous stem cell transplantation (arm B) compared with standard treatment without bortezomib (arm A). For allanalyzed chromosomal aberrations, progression-free survival (PFS) and overall survival (OS) were at least equal or superior in the bortezomib arm compared with the standard arm. Strikingly, patients with del(17p13) benefited the most from the bortezomib-containing treatment: the median PFS in arm A was 12.0 months and in arm B it was 26.2 months (P = .024); the 3 year-OS for arm A was 17% and for arm B it was 69% (P = .028). After multivariate analysis, del(17p13) was an independent predictor for PFS (P < .0001) and OS (P < .0001) in arm A, whereas no statistically significant effect on PFS (P = .28) or OS (P = .12) was seen in arm B. In conclusion, the adverse impact of del(17p13) on PFS and OS could be significantly reduced by bortezomib-based treatment, suggesting that long-term administration of bortezomib should be recommended for patients carrying del(17p13). This trial is registered at the International Standard Randomised Controlled Trial Number Register as ISRCTN64455289.
Introduction
Although significant progress has been made in the management of multiple myeloma (MM) patients, resulting in an improvement of survival (especially in younger patients), MM remains an incurable disease.1 The course of the disease shows heterogeneity, with widely diverging survival times from months to years. For this reason, prognostic factors are needed to determine the course of the disease, to define therapeutic strategies, and to predict long-term outcome. The combination of serum β2-microglobulin level with serum albumin concentration has been proposed as an outcome predictor in the International Staging System (ISS).2 Other prognostic parameters that are able to differentiate between high- and standard-risk diseases include lactate dehydrogenase levels, C-reactive protein levels, and proliferation based on plasma cell labeling.3 More recently, subgroups of MM have been defined by genetic and cytogenetic abnormalities and found to be associated with unique biologic, clinical, and prognostic features.4 FISH analysis on sorted CD138+ plasma cells can detect specific changes in interphase cells, overcoming the problem of the lack of dividing cells required for conventional cytogenetics. Abnormalities such as t(4;14), t(14;16), partial or whole chromosome 13 deletion, and loss of 17p13 carry a poor prognosis in patients undergoing high-dose therapy, whereas hyperdiploidy and t(11;14) translocations are associated with better outcomes.3,4
It is still a matter of debate whether novel drugs such as bortezomib or lenalidomide are able to improve outcome in patients with high-risk chromosomal aberrations. In particular, some previous studies suggested that bortezomib and lenalidomide are able to overcome the adverse effects associated with t(4;14) and del(13q14), but not those with del(17p13),5-8 whereas other studies show contradicting results.9,10 These apparently conflicting results might be because of small patient numbers, different therapeutic strategies, and the retrospective nature of some of these studies.
In the present study, we evaluated the association of high-risk FISH cytogenetics with the outcome of a subgroup of patients within the Stitching Hemato Oncologie voor Volwassenen Nederland (HOVON-65)/German Speaking Myeloma Multicenter Group (GMMG-HD4) trial, a prospective, randomized phase 3 trial for patients with newly diagnosed MM. Because only patients in treatment arm B received a bortezomib-based induction and maintenance therapy before and after high-dose chemotherapy, followed by autologous stem cell transplantation, we were able to analyze the therapeutic influence of bortezomib on chromosomal aberrations compared with patients receiving standard therapy in arm A.
Methods
Patients
A total of 833 patients (18-65 years of age) with newly diagnosed Salmon and Durie stage II-III MM were enrolled in a prospective, randomized phase 3 trial (HOVON-65/GMMG-HD4; EudraCT number 2004-000944-26) in 75 centers in the Netherlands, Germany, and Belgium. The German sites decided to perform a comprehensive FISH analysis, which is presented here. Patients with amyloidosis or monoclonal gammopathy of unknown significance and baseline peripheral neuropathy of grade 2 or more were excluded. The trial was done in accordance with the Declaration of Helsinki (Version 1996), and was approved by the local ethics committees of all participating institutions. We obtained written informed consent from the patients for treatment and sample procurement.
Patients were randomly assigned to arm A or arm B. Arm A consisted of 3 cycles of induction treatment with vincristine 0.4 mg IV on days 1-4; doxorubicin 9 mg/m2 IV on days 1-4; and dexamethasone 40 mg orally on days 1-4, 9-12, and 17-20. Arm B consisted of bortezomib 1.3 mg/m2 IV on days 1, 4, 8, and 11; doxorubicin 9 mg/m2 IV on days 1-4; and dexamethasone 40 mg orally on days 1-4, 9-12, and 17-20. Stem cells were mobilized by the use of cyclophosphamide 1000 mg/m2 IV on day 1, doxorubicin 15 mg/m2 IV on days 1-4, dexamethasone 40 mg orally on days 1-4, and G-CSF (filgrastim 10 μg/kg or lenograstim 300 μg/m2) per day subcutaneously divided into 2 doses per day from day 9 until the last stem cell collection. After stem cell collection, patients were treated with 1 or 2 cycles of high-dose melphalan (200 mg/m2 IV) and autologous stem cell rescue, followed by maintenance treatment with thalidomide (50 mg/d orally in arm A) or bortezomib (1.3 mg/m2 IV once every 2 weeks in arm B) for 2 years. For more detailed information, see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Follow-up data on PFS and OS were obtained up to April 12, 2011.
Enrichment of CD138+ plasma cells
Density gradient centrifugation of BM aspirates over Ficoll Hypaque (Biochrom) was performed to separate mononuclear cells by standard protocol. CD138+ plasma cells were isolated by MACS using anti-CD138 immunobeads and an auto-MACS separation system (Miltenyi Biotec) according to the manufacturer's protocol. Purity was confirmed by the CD38+ and CD138+ phenotypes in flow cytometric analysis.
Interphase FISH analyses
Interphase FISH analysis was accomplished on CD138-purified plasma cells as described previously11 using probes for the detection of numerical aberrations of the chromosome regions 1q21, 5p15/5q35 (only if necessary to define hyperdiploidy), 6q21, 8p21, 9q34, 11q23, 13q14.3, 15q22, 17p13, 19q13, and 22q11, as well as for the IgH translocations t(11;14)(q13;q32), t(4;14)(p16.3;q32), and t(14;16)(q32;q23). Hybridization was performed according to the manufacturer's instructions (Kreatech) and for the t(14;16) (Vysis). A total of 100 interphase nuclei per probe were evaluated using a DM RXA epifluorescence microscope (Leica). Hybridization efficiency was validated on interphase nuclei obtained from the peripheral blood and BM of a healthy donor. The thresholds for gains, deletions, and translocations were set at 10%. The score of Wuilleme et al was used to assess ploidy.12 Gains of at least 2 of the 3 chromosomes 5, 9, and 15 were used for a FISH definition of hyperdiploidy.
Statistical analysis
Frequency of cytogenetic abnormalities was assessed for imbalances between treatment arms using the Fisher exact test. PFS was defined as time from randomization until progression, relapse, or death, whichever came first. OS was calculated from randomization until death from any cause. Patients still alive were censored at the date of last contact. Estimation of PFS and OS distribution was performed by the method of Kaplan and Meier. The log-rank test was used for comparisons of OS and PFS curves. Univariate and multivariate Cox proportional hazards (PH) regression analysis was used to evaluate the prognostic impact based on hazard ratios (HRs) including 95% confidence intervals (95% CIs). Multiple imputations using predictive mean matching were performed for the multivariate analysis. The P values of the univariate Cox PH regression were adjusted for multiple testing using the Bonferroni-Holm correction. P < .05 was considered statistically significant. All statistical computations were carried out with R Version 2.12.0 statistical software using the add-on R package Hmisc.
Results
Patient cohort
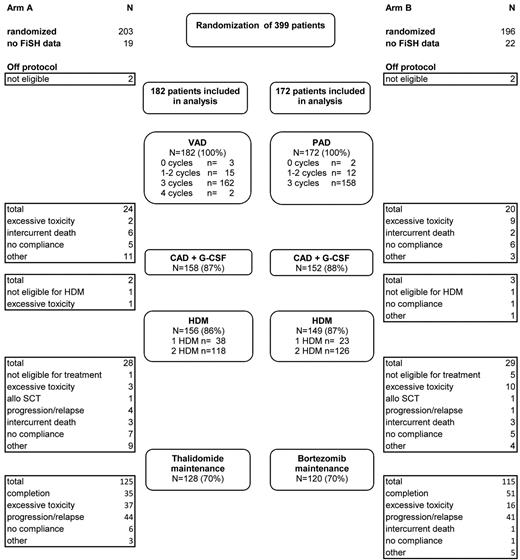
BM aspirates from 354 of 395 eligible patients treated at 35 different institutions in Germany were sent to a central laboratory in Heidelberg and analyzed for chromosomal aberrations by FISH (Figure 1). The median follow-up time for all patients from randomization was 40.9 months (95% CI, 39.7-42.5). The median age of the patients was 57 years (range, 25-65 years). The distribution of ISS stages was as follows: stage I, 37.0%; stage II, 32.5%; and stage III, 23.4% (missing data, 7.1%). Patients treated in arm A (n = 182) or arm B (n = 172) displayed similar median age, gender ratio, isotype repartition, and distribution of ISS stages. For the entire group, the median PFS time was 33.4 months; the median OS time was not yet reached. Patients randomized to arm B had a somewhat longer PFS (median PFS 35.7 vs 31.2 months; HR = 0.80; 95% CI, 0.61-1.06; P = .12) and OS (3-year OS rate 84% vs 73%; HR = 0.65; 95% CI, 0.43-1.01; P = .053) compared with patients treated in arm A.
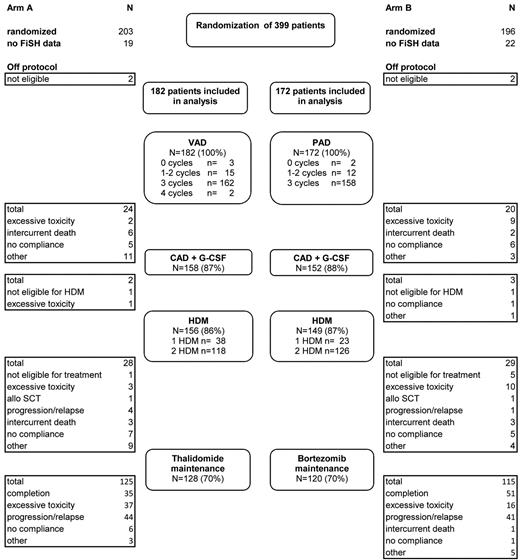
Diagram of patient disposition and patient flow through protocol. VAD indicates vincristine plus doxorubicin plus dexamethasone; PAD, bortezomib plus doxorubicin plus dexamethasone; CAD, cyclophosphamide plus doxorubicin plus dexamethasone; and HDM, high-dose melphalan followed by autologous stem cell transplantation.
Diagram of patient disposition and patient flow through protocol. VAD indicates vincristine plus doxorubicin plus dexamethasone; PAD, bortezomib plus doxorubicin plus dexamethasone; CAD, cyclophosphamide plus doxorubicin plus dexamethasone; and HDM, high-dose melphalan followed by autologous stem cell transplantation.
Frequencies of chromosomal aberrations
Chromosomal aberrations were detected in 341 of 354 (97%) patients. Interphase FISH analysis on CD138-enriched plasma cells revealed gains of chromosome regions 1q21 (32.3%), 9q34 (57.0%), 11q23 (48.6%), 15q22 (52.2%), and 19q13 (52.0%), as well as deletions of chromosome regions 8p21 (23.3%), 13q14 (48.3%), and 17p13 (10.6%), as shown in Table 1. Furthermore, the IgH translocations t(11;14), t(4;14), and t(14;16) were observed at a frequency of 19.2%, 14.2%, and 1.8%, respectively. Applying the score by Wuilleme et al, hyperdiploidy was found in 49.4% of MM patients.12 The distribution of chromosomal aberrations analyzed was similar in both treatment arms.
Correlation of chromosomal aberrations with patient outcome
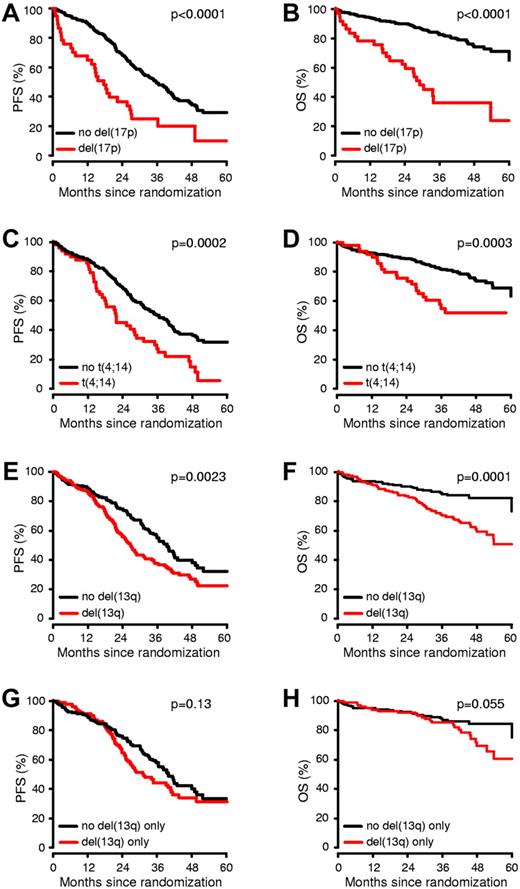
We analyzed the prognostic impact of chromosomal aberrations on PFS and OS (Figure 2 and Table 1). The presence of del(13q14), del(17p13), t(4;14), and +1q21 had a significant adverse impact on both PFS and OS. In addition, patients with +19q13 displayed a favorable OS, but this effect disappeared after adjustment of P values for multiple testing.
Impact of del(17p13), t(4;14) and del(13q14) on PFS and OS. MM patients were stratified by the presence or absence of each of the specific cytogenetic abnormalities showing statistical significance in the univariate analysis (A-F). In patients lacking t(4;14) and del(17p13), del(13q14) was no longer prognostic (G-H).
Impact of del(17p13), t(4;14) and del(13q14) on PFS and OS. MM patients were stratified by the presence or absence of each of the specific cytogenetic abnormalities showing statistical significance in the univariate analysis (A-F). In patients lacking t(4;14) and del(17p13), del(13q14) was no longer prognostic (G-H).
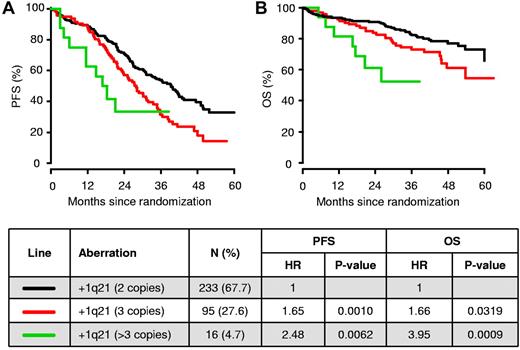
The median PFS time for +1q21 was 26.6 months, compared with 39.3 months for patients lacking this abnormality (HR = 1.7; P = .0002). Similar results were observed for OS with 3-year OS rates of 70% vs 82% (HR = 1.9; P = .0052), respectively. Of all patients analyzed with +1q21, we identified 16 patients with> 3 copies of this chromosomal region (Figure 3). Compared with patients with a normal copy number of 1q21 (2 copies), the median PFS times for patients carrying 3 or > 3 copies of 1q21 were 28.0 months (HR = 1.7; P = .0010) and 17.6 months (HR = 2.5; P = .0062), respectively. Similar results were observed for OS: the probabilities of OS at 3 years decreased from 73% (HR = 1.7; P = .032) to 52% (HR = 4.0; P = .0009) in patients carrying 3 or > 3 copies of 1q21, respectively.
PFS and OS among MM patients according to the copy numbers of +1q21. Kaplan-Meier analysis of PFS (A) and OS (B) is displayed in relation to no +1q21 (2 copies of 1q21; n = 233), +1q21 (3 copies; n = 95), and +1q21 (> 3 copies; n = 16) in patients treated within the HOVON-65/GMMG-HD4 trial.
PFS and OS among MM patients according to the copy numbers of +1q21. Kaplan-Meier analysis of PFS (A) and OS (B) is displayed in relation to no +1q21 (2 copies of 1q21; n = 233), +1q21 (3 copies; n = 95), and +1q21 (> 3 copies; n = 16) in patients treated within the HOVON-65/GMMG-HD4 trial.
Of all of the analyzed chromosomal aberrations, del(17p13) showed the most profound effect on outcome. Median PFS time was 17.6 months for patients with del(17p13) compared with 35.7 months for patients lacking this abnormality (HR = 2.5; P < .0001). Similar results were observed for OS, with 3-year OS rates of 36% versus 83% (HR = 4.4; P < .0001), respectively.
Patients with t(4;14) showed a significantly worse median PFS time (21.7 vs 35.7 months; HR = 2.0; P = .0002) and 3-year OS rate (55% vs 82%; HR = 2.4, P = .0003) compared with patients lacking this aberration.
The median PFS time for patients with del(13q14) was 26.6 months compared with 39.3 months for those without (HR = 1.5; P = .0023); the 3-year OS for patients carrying del(13q14) was 70% compared with 85% for those without the deletion (HR = 2.4; P = .0001). The presence of del(13q14) was positively correlated with del(17p13) (P < .0001) and t(4;14) (P < .0001). In patients lacking t(4;14) and del(17p13), del(13q14) was no longer of prognostic significance.
ISS- and FISH-based prognostication scheme
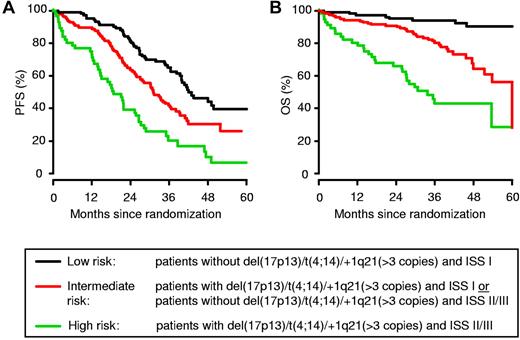
Based on the results in the univariate analysis, we analyzed whether combining the ISS score with information on the presence of high-risk aberrations such as del(17p13), t(4;14), or +1q21 (> 3 copies) could improve the prognostic value with regard to patient outcome (Figure 4 and Table 2). A combination of the presence or absence of del(17p13), t(4;14), or +1q21 (> 3 copies) with the ISS score allowed patients to be stratified into 3 distinct groups: (1) low-risk, including patients with the absence of del(17p13)/t(4;14)/1q21 (> 3 copies) and an ISS score of I]; high-risk, patients with the presence of del(17p13)/t(4;14)/1q21 (> 3 copies) and an ISS score of II or III; and (3) intermediate-risk, including all remaining patients. Most of the patients belonged to the low-risk (33%) and intermediate-risk (49%) groups, whereas 18% were allocated to the high-risk group. The median PFS times for the low-, intermediate-, and high-risk groups were 41.9 months, 31.1 months (HR = 1.7; P = .0018), and 18.7 months (HR = 3.6; P < .0001), respectively. The 3-year OS decreased from 94% in the low-risk group to 80% (HR = 4.6; P = .0001) and 43% (HR = 12.8; P < .0001) in the intermediate- and high-risk groups, respectively.
Combining information on chromosomal aberrations del(17p13), t(4;14), and +1q21 (> 3 copies) with ISS score allows stratification of MM patients undergoing high-dose chemotherapy followed by autologous SCT. The combination of the presence or absence of del(17p13), t(4;14), and +1q21 (> 3 copies) with the ISS score allowed stratification of patients into 3 distinct groups: low-risk, high-risk, and intermediate-risk (all remaining patients), representing 33%, 49%, and 18% of patients, respectively.
Combining information on chromosomal aberrations del(17p13), t(4;14), and +1q21 (> 3 copies) with ISS score allows stratification of MM patients undergoing high-dose chemotherapy followed by autologous SCT. The combination of the presence or absence of del(17p13), t(4;14), and +1q21 (> 3 copies) with the ISS score allowed stratification of patients into 3 distinct groups: low-risk, high-risk, and intermediate-risk (all remaining patients), representing 33%, 49%, and 18% of patients, respectively.
Comparison between treatment arms
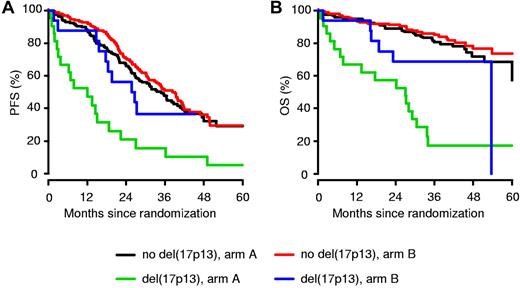
For all analyzed chromosomal aberrations, the median PFS times and 3-year OS rates were at least equal or superior in the bortezomib arm compared with the standard arm (Table 3). However, a statistically significant difference was found only for patients carrying del(17p13). Patients with del(17p13) in arm B displayed a significantly better median PFS time compared with patients treated in arm A (26.2 vs 12.0 months; P = .024; Figure 5). Moreover, bortezomib-based treatment resulted in an improved 3-year OS rate for patients with del(17p13) (17% for arm A vs 69% for arm B; P = .028), whereas the 3-year OS was 80% and 85% (P = .41), respectively, in patients without del(17p13). For del(17p13), different thresholds of plasma cells presenting the abnormality were analyzed and found to be correlated with patient outcome. The survival benefit for patients with del(17p13) receiving bortezomib-based treatment was confirmed when a cut-off of 60% was used to define the abnormality (median PFS time for arm A was 12.0 months and for arm B, 25.7 months; P = .017; the 3-year OS rate was 8% for arm A and 62% for arm B; P = .037).
Impact of del(17p13) on PFS and OS. For all patients with del(17p13), the median PFS times (A) and 3-year OS rates (B) in the bortezomib-based treatment arm B were better compared with the standard arm A.
Impact of del(17p13) on PFS and OS. For all patients with del(17p13), the median PFS times (A) and 3-year OS rates (B) in the bortezomib-based treatment arm B were better compared with the standard arm A.
Multivariate analysis
Because del(13q14) was strongly correlated with the presence of t(4;14) and del(17p13), the prognostic value of del(13q14) was analyzed without concurrent t(4;14) and del(17p13) in a Cox PH model (Table 4). For the entire group of patients, del(17p13), +1q21 (3 copies), +1q21 (>3 copies), and ISS stage III were identified as independent predictors for adverse PFS and OS. In addition, treatment arm B and +19q13 were associated with improved OS. When the treatment arms were analyzed separately, del(17p13) was found to be an independent predictor for PFS (HR = 4.13; P < .0001) and OS (HR = 6.71; P < .0001) in arm A. In contrast, in arm B (the bortezomib arm), the presence of del(17p13) was not associated with a statistically significant effect on PFS (HR = 1.56; P = .28) or OS (HR = 2.60; P = .12).
Discussion
Our analysis, which was based on the results of a prospective, randomized phase 3 trial, was performed to evaluate the prognostic and predictive value of genomic aberrations in patients with newly diagnosed MM. Our results show that, beyond global cytogenetic risk classification, chromosomal aberrations are clinically significant factors with a treatment-associated prognostic value that can be modified by the introduction of a novel drug such as bortezomib. For molecular cytogenetic analysis, BM specimens were sent from 35 different sites to a central laboratory, showing that genetic information can be obtained in approximately 97% of patients even in a multicenter setting. All interphase FISH studies were performed on CD138-enriched plasma cells, which were analyzed with a comprehensive set of 12 different DNA probes specific for the most recurrent chromosomal aberrations observed in MM. The frequencies of chromosomal aberrations were consistent with those in previous studies.11,13,14 For the entire group of patients, we found that del(17p13), del(13q14), t(4;14), and +1q21 were linked to poor outcome, as shown for both PFS and OS in the univariate analysis. After multivariate analysis, del(17p13), +1q21 (3 copies), +1q21 (> 3 copies), and ISS stage III were identified as independent predictors for adverse PFS and OS.
Although gains of chromosome 1q were proposed as an adverse prognostic factor in previous studies,15,16 we show herein that the outcome of patients is related to the copy number variation of +1q21 found in MM cells. We identified 16 patients (4.7%) with more than 3 copies of this chromosomal region, and only 1 of these patients displayed a concurrent del(17p13). The clinical course of these patients with > 3 copies of +1q21 was characterized by a short median PFS time of 17.6 months and a 3-year OS rate of 52%, whereas exactly 3 copies of +1q21 was associated with only a marginal effect on outcome. This finding suggests that increased copy numbers of +1q21 are linked with adverse outcome, possibly because of a dosage effect of genes located at this chromosomal region. In agreement with this finding, recent expression profiling data confirm the critical role of genes located on chromosome 1 in the survival of MM patients.17 Shaughnessy et al investigated the gene expression profile of 532 newly diagnosed MM patients and identified 70 genes linked to early disease-related death. Strikingly, 30% of these genes were located on chromosome 1, with most of the down-regulated genes located on the short arm of chromosome 1 and most of the up-regulated genes on 1q. In particular, increased copy numbers of CKS1B and the IL-6 receptor mapping within a minimally amplified region of chromosome 1q21 were found to be correlated with poor outcome in MM.18,19
Using univariate analysis, we confirmed that t(4;14) is an important prognostic factor for outcome in MM. Interestingly, t(4;14) was only of marginal significance for PFS and OS in the multivariate analysis. The reason for this might be that the prognostic information of chromosomal aberrations was analyzed together with the ISS score in our statistical model. The French study group Intergroupe Francophone du Myélome (IFM) showed that the outcome of patients with t(4;14) and low β2-microglobulin levels of ≤ 4 mg/L was similar to that of patients without the translocation but with high β2-microglobulin levels of > 4 mg/L.14 In addition, we found that +1q21 was strongly correlated with the presence of t(4;14). Therefore, t(4;14) loses some of its prognostic power when it is analyzed together with +1q21 in the same statistical model.
Although del(13q14) was confirmed as a powerful prognostic marker by univariate analysis, a more detailed analysis showed that most of the prognostic power of del(13q14) was related to t(4;14) and del(17p13), which are frequently associated with del(13q14). In patients lacking t(4;14) and del(17p13), del(13q14) was no longer prognostic, confirming previously published data.13,14 Therefore, the prognostic value of del(13q14) was analyzed without concurrent t(4;14) and del(17p13) in our multivariate analysis.
Recently, we and Avet-Loiseau et al reported that combining the ISS score with information on the presence of high-risk aberrations can improve prognostic value with regard to MM patient outcome.11,20 To our previous publication, we added +1q21 (> 3 copies) to the group of high-risk aberrations, because the outcome of these patients was almost as low as it was observed for patients with del(17p13). We developed an ISS- and FISH-based prognostication scheme, which allows a risk stratification of patients treated in the HOVON-65/GMMG-HD4 trial into 3 groups: a low-risk group, a high-risk group, and all others. Although the results obtained using this model appear to be very robust, they need to be confirmed in independent patient cohorts.
Because of the 2-arm design of the HOVON-65/GMMG-HD4 trial, we were able to analyze the effect of bortezomib on cytogenetically defined subgroups of patients. In all subgroups, PFS and OS were at least equal or superior in the bortezomib arm compared with the standard arm. Strikingly, patients with del(17p13) benefited the most from the bortezomib-containing treatment. After multivariate analysis, del(17p13) was an independent predictor for PFS and OS in arm A, whereas no statistically significant effect on PFS and OS was seen in arm B. Patients in the standard arm of our study received a maintenance therapy with thalidomide and, based on the experience of the MRC Myeloma IX trial,21 thalidomide maintenance therapy is associated with a significantly longer PFS time than no maintenance (23 vs 15 months; P < .001). However, the use of thalidomide maintenance was associated with PFS benefits and a potential OS benefit only in patients with favorable FISH, but worse OS in patients with adverse FISH as defined by the presence of the cytogenetic abnormalities gain(1q), t(4;14), t(14;16), t(14;20), and del(17p). In the present study, patients receiving maintenance with thalidomide displayed a median PFS time of 12 months, which is slightly worse than that observed in 2 other studies finding median event-free survival times of 14 and 15 months for patients with del(17p13) in predominantly thalidomide-naive patients.7,14 Therefore, we cannot exclude that the beneficial effect of bortezomib in patients with deletion 17p13 is overestimated when thalidomide is used in the control arm.
Although patients with del(17p13) showed an improved outcome in our study when treated with bortezomib, the prognosis was still inferior compared with patients without this cytogenetic aberration, suggesting that long-term administration of bortezomib is able to improve, but not to fully overcome, the adverse outcome associated with del(17p13). Our data are in agreement with the conclusion drawn from the Arkansas experience that incorporation of bortezomib in the Total Therapy 3 protocol negated the adverse consequences of del(17p13), at least in MM patients who belonged to the low-risk group as defined by gene-expression profiling.10 However, induction with 4 cycles of bortezomib and dexamethasone without further administration of bortezomib in the consolidation or maintenance phase did not show an advantage to vincristine plus doxorubicin plus dexamethasone induction in the Intergroupe Francophone du Myélome (IFM) experience for patients with del(17p13).7 Therefore, our randomized study supports the idea that long-term administration of bortezomib is required to improve the outcome of patients carrying del(17p13), because patients in the HOVON-65/GMMG-HD4 and Total Therapy 3 trials received 64 and almost 150 injections of bortezomib per protocol, respectively, whereas only 16 injections of bortezomib were administered in the IFM study.7,10 But why is the effect seen for bortezomib-treated patients on del(17p13) more profound than on any other chromosomal aberration in our study? It is widely accepted that the relevant target of del(17p13) is the tumor suppressor TP53, because low TP53 gene expression is strongly correlated with 17p13 deletion.22 Initially, the molecular mechanisms of proteasome inhibition mediating antimyeloma activity were proposed to depend on the abrogation of the NF-κB pathway and the activation of p53-induced downstream effector molecules.23,24 However, Hideshima et al recently showed that bortezomib-induced cytotoxicity cannot be fully attributed to inhibition of canonical NF-κB activity in myeloma cells.25 Furthermore, evidence has been accumulating during recent years that protein homeostasis represents the major “Achilles heel” of myeloma cells and that proteasome inhibition profoundly perturbs this sensitive balance of intracellular protein production and disposal.26-28 Therefore, our data strongly support the concept that high levels of immunoglobulin production and endoplasmic reticulum–Golgi protein transport sensitize myeloma cells to proteotoxic stress and that bortezomib induces apoptosis in these tumor cells by a p53-independent mechanism.29
Recently, the IFM reported that bortezomib-based treatment is able to improve outcome in myeloma patients with t(4;14).7 Concordantly, we found that patients with t(4;14) receiving bortezomib-based treatment displayed a prolonged median PFS time (25.3 vs 21.7 months) and improved 3-year OS rate (66% vs 44%) compared with patients treated in the standard arm. However, this effect was not as profound as that shown in the retrospective analysis by the IFM. This might be because of some differences in the treatment protocols, explaining especially the better results for patients with t(4;14) treated in the standard arm of the HOVON-65/GMMG-HD4 trial (median PFS time 21.7 months) compared with French series (median event-free survival time, 16 months).
Bortezomib and lenalidomide were successfully combined in the upfront setting (lenalidomide, bortezomib, and dexamethasone) with high-quality results seen in terms of overall quality of depth and duration of response.30 Moreover, whereas sample size (n = 66) limits conclusions in any phase 1/2 study, it was intriguing to see in this study that the presence of adverse cytogenetics including t(4;14) and del(17p13) was not associated with worse outcome.
In conclusion, the considerable prognostic implications of the analyzed chromosomal aberrations confirm and substantially extend the results of previous studies.5-10,13,14 Our data show that bortezomib-based treatment is able to improve outcome in patients with MM, including patients with high-risk chromosomal aberrations. Therefore, we support the concept of the early use of novel agents such as bortezomib and lenalidomide in MM patients with high-risk disease.31 We and others have shown that bortezomib and lenalidomide are active in MM, including patients with high-risk chromosomal aberrations such as del(17p13) and t(4;14).7,32 In addition, thalidomide seems to be a treatment option for patient with standard-risk cytogenetics.21
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Steffen Luntz and Bärbel Schurich from the Coordination Center for Clinical Trials at the University of Heidelberg for excellent administrative support; Katrin Heimlich, Maria Dörner, and Hildegard Bethäuser for technical assistance in the enrichment of CD138+ plasma cells; and Michaela Brough, Stephanie Pschowski-Zuck, and Desireé Kirn for performing interphase FISH analysis.
The HOVON-65/GMMG-HD4 trial (EudraCT number 2004-000944-26) was supported by the Dutch Cancer Foundation, by the German Federal Ministry of Education and Research, and by a grant from Janssen Cilag. The GMMG also received grants for this trial by Novartis, Amgen, Chugai, Roche, and the Tumorzentrum Heidelberg/Mannheim.
Authorship
Contribution: P.S. and H.G. conceived and designed the study; H.v.d.V. provided financial support; U.B. provided administrative support; K.N., D.H., H.S., I.W.G., M.P., U.D., K.W., H.M. W.L., C.T., M.H., C.S., H.G.D., N.P., I.G.H.S.-W., M.S.R., A.D.H., A.J., and H.G. provided study materials or patients; K.N., U.B., T.H., B.v.d.H., D.H., M.S.R., A.J., H.M.L., P.S., and H.G. collected and assembled the data; K.N., T.H., D.H., M.S.R., A.J., and H.G. analyzed and interpreted the data; K.N., U.B., T.H., D.H., M.S.R., A.J., and H.G. wrote the manuscript; and all authors gave final approval of the manuscript.
Conflict-of-interest disclosure: K.N. has received honoraria from Celgene, Janssen-Cilag, and Novartis and served on the advisory board for Celgene and Janssen-Cilag; H.L. has been a consultant for Celgene and Genmab; H.S. has received honoraria from and served on the advisory board for Celgene and Janssen-Cilag; K.W. has received honoraria from Celgene and Janssen-Cilag and served on the advisory board for Celgene; M.P. served on the advisory board for Celgene and Onyx; C.S. served on the advisory board for Celgene, Janssen-Cilag, and Novartis; I.S.W. has received honoraria from and served on the advisory board for Celgene and Janssen-Cilag; H.V. is an employee of the pharmaceutical industry (Janssen Research & Development) and has stock ownership interest in Johnson & Johnson; P.S. has received research support from Celgene, Janssen-Cilag, and Onyx and served on the advisory board for Celgene, Janssen-Cilag, Millennium, and Onyx; H.G. has received research support from Celgene, Janssen-Cilag, Novartis, Chugai, and Roche and served on the advisory board for Celgene and Janssen-Cilag. The remaining authors declare no competing financial interests.
Correspondence: Kai Neben, MD, Department of Internal Medicine V, University of Heidelberg, Im Neuenheimer Feld 410, 69120 Heidelberg, Germany; e-mail: kai.neben@med.uni-heidelberg.de.