Abstract
We investigated the therapeutic potential of JQ1, an inhibitor of the BET class of human bromodomain proteins, in B-cell acute lymphoblastic leukemia (B-ALL). We show that JQ1 potently reduces the viability of B-ALL cell lines with high-risk cytogenetics. Among the most sensitive were lines with rearrangements of CRLF2, which is overexpressed in ∼ 10% of B-ALL. CRLF2 heterodimerizes with the IL7 receptor (IL7R) and signals through JAK2, JAK1, and STAT5 to drive proliferation and suppress apoptosis. As previously observed, JQ1 induced the down-regulation of MYC transcription, the loss of BRD4 at the MYC promoter, and the reduced expression of c-Myc target genes. Strikingly, JQ1 also down-regulated IL7R transcription, depleted BRD4 from the IL7R promoter, and reduced JAK2 and STAT5 phosphorylation. Genome-wide expression profiling demonstrated a restricted effect of JQ1 on transcription, with MYC and IL7R being among the most down-regulated genes. Indeed, IL7R was the only cytokine receptor in CRLF2-rearranged B-ALL cells significantly down-regulated by JQ1 treatment. In mice xenografted with primary human CRLF2-rearranged B-ALL, JQ1 suppressed c-Myc expression and STAT5 phosphorylation and significantly prolonged survival. Thus, bromodomain inhibition is a promising therapeutic strategy for B-ALL as well as other conditions dependent on IL7R signaling.
Introduction
Long-term disease-free survival is achieved in > 80% of children with B-cell acute lymphoblastic leukemia (B-ALL) but only 40% of adults. Particular genomic alterations in B-ALL, including mutations in the transcription factor IKZF1 and rearrangements of both MLL and the cytokine receptor subunit CRLF2 confer a poor prognosis in both children and adults.1-6 In addition, current therapies are associated with significant short- and long-term toxicities. Newer agents that target specific deregulated oncogenic pathways are currently in clinical trials, and their success coupled with advances in the genetic characterization of B-ALL augurs a new era of pharmacogenetic therapies.7,8
We and others have recently described the therapeutic potential of pharmacologic inhibition of bromodomains in hematologic malignancies.9-12 Bromodomains mediate noncovalent protein-protein interactions by associating with posttranslationally acetylated lysine residues.13 Most of these recent studies used JQ1, a novel thieno-triazolo-1,4-diazapine that binds selectively and with high affinity to the acetyllysine binding pocket of the conserved bromodomain and extra-terminal domain (BET) protein family (BRD2, BRD3, BRD4, and BRDT).14
A major function of the BET bromodomain BRD4 is the recognition of acetylated histones found in transcriptionally active regions of chromatin, which promotes the recruitment of transcriptional activators.15 BRD3 and BRD4 are known to drive oncogenesis when translocated with the NUT gene in a rare form of squamous carcinoma.16 The first biologic validation of BRD4 as a therapeutic target for hematologic neoplasia was established by the laboratories of Vakoc and Lowe.9 Using an elegant chromatin factor-focused RNAi screen in a mouse model of MLL-induced AML, they found that specific knockdown of murine Brd4 significantly decreased disease progression. Pharmacologic inhibition of bromodomains in this model and others is thought to selectively target malignant cells by disrupting chromatin-mediated signal transduction and thus reducing transcription at oncogene loci, most notably MYC, which encodes the protein c-Myc.9-12
In this study, we used both cell line and in vivo models to determine the translational potential of BET bromodomain inhibition in subtypes of B-ALL. Our findings indicate that not only does BET inhibition result in a potent suppression of MYC transcription and activity but also dramatically decreases expression of the cytokine receptor IL7R in CRLF2-rearranged and other B-ALL cell types.
Methods
Reagents
B-ALL cell lines used were NALM-6,17 MHH-CALL4,18 MUTZ-5,19 CEMO-1,20 Reh,21 697,22 RS4;11,23 and SEMK2,24 as well as the chronic myelogenous leukemia line K562.25 Cell lines were grown in RPMI 1640 supplemented with 10% FBS (or 20% for MHH-CALL4, MUTZ-5, 697, and CEMO-1), 2mM l-glutamine, penicillin (100 U/mL), and streptomycin (50 μg/mL), and were grown at 37°C in 5% CO2. JQ1 was synthesized as previously described14 and diluted in DMSO for all cellular assays.
Cell viability, proliferation, and caspase activity assays
For dose-response cellular viability assays, cells were seeded onto 384-well tissue culture-treated plates at a density of 2.5 × 104 cells/well (MHH-CALL4 and MUTZ-5) or 5 × 103 cells/well (CEMO-1, Reh, NALM-6, 697, K562, RS4;11, SEMK2) in a volume of 50 μL/well. Addition of JQ1 was performed with a JANUS Workstation (PerkinElmer Life and Analytical Sciences) using a 384-well pinhead tool that is calibrated to deliver 100 nL drug/well. After 72 hours of incubation with JQ1, cells were analyzed for cell viability using the CellTiterGlo (Promega) luminescent assay kit per the manufacturer's instructions. Luminescence was read on an EnVision 2104 Multilabel Plate Reader (PerkinElmer Life and Analytical Sciences). Nonlinear dose-response curves were fitted to the data using Graphpad Prism Version 5.04 software. From these curves, GI50 values (the concentration of JQ1 at which the fraction of affected cells is 50%) and Emax (the fraction of affected cells at the maximum effect of JQ1). For time-course assays, cells were cultured in tissue culture flasks at a starting density of ∼ 5 × 105 cells/mL and treated with 500nM JQ1 or vehicle (DMSO, < 0.1%). Samples were drawn, counted using a standard hemacytometer, and plated onto 384-well plates. Viability was measured using CellTiterGlo, and caspase activity was measured using the CaspaseGlo 3/7 assay kit (Promega) per the manufacturer's instructions.
Flow cytometric studies
For quantifying cell death, cells (1 × 106/mL) were treated with JQ1 (500nM) or vehicle (DMSO 0.1%). The cells were washed and resuspended in annexin V/propidium iodide buffer solution (10mM HEPES pH 7.4, 140mM NaCl, 2.5mM CaCl2) containing annexin V–FITC (BD Biosciences PharMingen, 51-66211e) and propidium iodide (BD Biosciences Pharmingen, 51-65874x). For cell cycle analyses, cells (1 × 106/mL) were treated with JQ1 (500nM) or vehicle (DMSO 0.1%). Cells were washed with PBS, fixed with 70% ethanol overnight, and washed with PBS. RNA was degraded with RNase (Roche Diagnostics, 10109169001), and DNA was stained with propidium iodide. For detection of CRLF2 and IL7 receptor (IL7R) expression, cells (1 × 106/mL) were treated with drug and vehicle, washed once in PBS before and after double staining with hCD127-ALEXA 647 (IL7R; BD Biosciences, 558598) and hTSLPR (CRLF2)–PE (eBioscience, 12-5499-73). An IgG isotype antibody was used as a negative control: for IL7R flow, IgG (BD Biosciences, 557783); and for CRLF to flow, IgG (eBioscience, 12-4724-42). All samples were analyzed on a BD FACSCanto II. Histograms were generated and cell cycle analysis was performed using FlowJo Version 7.6.1 software (TreeStar).
Expression analysis
Cells (5 × 106) were treated with 500nM JQ1 or DMSO (< 0.2%) in a 6-well dish, and cells were harvested at various time points. RNA extraction was performed with TRIzol reagent (Invitrogen, 15596-026). cDNA was made with Applied Biosciences reverse transcription reagents (ABI N808-0234), and quantitative PCR was carried out using an ABI 7300 real-time PCR system using PowerSYBR Green reagent (ABI 4367659) and the following primer sets (5′-3′): CRLF2 (forward) AACGCCTCCCAAACCAAAG, (reverse) GCACGCTGGGAATGAGAAA; MYC (forward) TCAAGAGGTGCCACGTCTCC, (reverse) TCTTGGCAGCAGGATAGTCCTT; IL7R (forward) CGCCAGGAAAAGGATGAAA, (reverse) ATACATTGCTGCCGGTTGG; β-actin (forward) ACCGAGCGCGGCTACAG, (reverse) CTTAATGTCACGCACGATTTCC; and GAPDH (forward) CCACTCCTCCACCTTTGAC, (reverse) ACCCTGTTGCTGTAGCCA.
Data for each time point were normalized to GAPDH expression relative to vehicle-treated cells and shown as a fold change to baseline expression at time = 0 hours.
For the c-Myc signature multiplexed expression analysis assays, a NanoString nCounter instrument was used (NanoString Technologies). Extracted RNA was added to NanoString reagents per the manufacturer's instructions. c-Myc target genes were culled from a c-MYC target gene database (www.myccancergene.org). All counts were normalized to the average of 3 housekeeping genes (GAPDH, ACTB, and TUBB). For each cell line, gene values were scaled to the gene's average expression value across samples. Data are represented with a heat map scale: blue represents a decrease in fold change relative to global normalization; white, no change; and red, an increase. Combined data across both MHH-CALL4 and MUTZ-5 cells lines were analyzed in the same manner, and unsupervised hierarchical clustering was performed by Euclidean distance of the unweighted average values with Spotfire DecisionSite Version 9.1.1 software.
ChIP
ChIP studies were carried out as described previously using specific primers for the MYC and IL7R locus.10 Primers were designed to amplify 2 sites within each promoter region (5′-3′), and a negative control region (NR) outside the promoter region: MYC promoter site 1 (forward) ACACTAACATCCCACGCTCTG and (reverse) GATCAAGAGTCCCAGGGAGA; MYC promoter site 2 (forward) GGTCGGACATTCCTGCTTTA and (reverse) GATATGCGGTCCCTACTCCA; MYC NR (forward) TCCTGGGTAGGAACCAGTTG and (reverse) ACTCACCAAGAGCTCCTCCA; IL7R promoter site 1 (forward) CTTCCTGTTTCTGGAACTTGC and (reverse) AGGGAGGGAGGAAGACAGAG; IL7R promoter site 2 (forward) CTTGTCAGGAGATCAAACTGGA and (reverse) GCACTGGTCACTGAAGACAAAG; and IL7R NR (forward) CGAGACACCAGCCCAGCGTG and (reverse) CTGGGTGAGTGCTTGGCGGG.
Enrichment data were analyzed by calculating the immunoprecipitated DNA percentage of input DNA for each sample.
Immunoblotting
Cells (1 × 106/mL) were treated with JQ1 (500nM) or vehicle (DMSO 0.1%), lysed, and protein was detected by Western blotting as described.10,26 For poly(ADP)–ribose polymerase (PARP), we used Cell Signaling #9542; for LC3 detection, we used Novus Biologicals #NB600-1384, and chloroquine from Sigma-Aldrich.
Gene expression arrays
Cells were treated with 500nM JQ1 or vehicle control (DMSO, > 0.1%). RNA was harvested as described under “Expression analysis.” and processed for oligonucleotide microarray profiling. Affymetrix Human Exon 1.0 ST arrays (GEO #GSE39995) were processed using the Affymetrix Expression Console RMA gene-level analysis, and batch corrected by cell type using ComBat.27 Probe sets with RMA values below negative control probes were excluded from analysis. P values for differential expression were determined and heatmaps generated using Spotfire DecisionSite. Gene sets were downloaded from the Broad Institute's MSigDB website.28 Gene set permutations were used to determine statistical enrichment of the gene sets in JQ1-treated versus DMSO-treated cells.
In vivo xenograft studies
The primary human B-ALL 537 bone marrow sample was xenotransplanted into irradiated NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice as previously described.26 Sample 537 harbors a P2RY8-CRLF2 rearrangement and lacks a somatic mutation within the known components of CRLF2 signaling, including IL7R, CRLF2, TSLP, JAK1, JAK2, and STAT5A/B. Eight-week-old female mice underwent secondary transplantation, and sentinel mice were monitored for engraftment. Once bone marrow involvement exceeded 30% (based on flow cytometry for hCD45 (BD Biosciences Pharmingen, 347463), xenografted mice were randomized to receive either JQ1 (50 mg/kg intraperitoneally daily), or vehicle (10% DMSO in D5W intraperitoneally daily). Mice were killed when they developed hind limb paralysis or became moribund. To assess pharmacodynamic endpoints, a separate cohort of mice was analyzed after 5 days of treatment. Spleens were fixed in 10% neutral-buffered formalin for immunohistochemistry. Staining for pSTAT5 was performed as previously described.29 Immunohistochemistry for c-Myc was performed with a c-Myc specific antibody (Epitomix, Y69) at 1:1000 with TSA (PerkinElmer Life and Analytical Sciences) amplification.30 Slides were subsequently analyzed using a Axio Observer.A1 (Zeiss) microscope AxioVision Rel. 4.7 (Zeiss) software. Images were acquired at 100× magnification. Complete blood counts were determined using a blood analyzer (Sysmex XT-2000i). Leukemic cells were also harvested from spleen samples for IL7R flow cytometry and RT-PCR. All animal studies were performed under Dana-Farber Cancer Center Animal Care and Use Committee approved protocols.
Results
Antiproliferative and proapoptotic effects of JQ1 in B-ALL cells
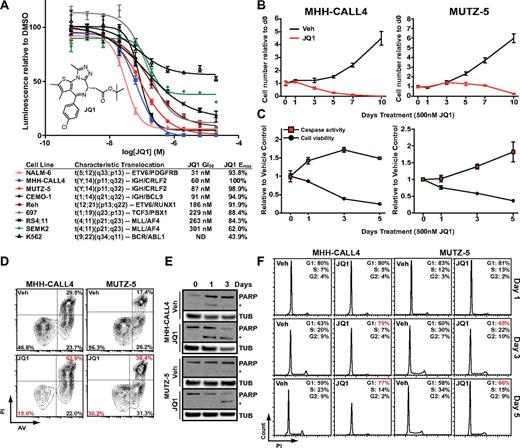
We first determined the effects of JQ1 on the proliferation of B-ALL cell lines with diverse chromosomal aberrations (Figure 1A). The BCR/ABL1-positive K562 chronic myelogenous leukemia line, previously shown to be relatively resistant to JQ1 treatment, was included as a control.9 JQ1 had marked effects on each B-ALL cell type, with 50% growth inhibition (GI50) values between 31 and 301nM. Of note, JQ1 had similar potency in the MLL-rearranged RS4;11 cell line (GI50 263nM) to the bromodomain inhibitor I-BET151 in a previous report12 (GI50 192nM; Figure 1A). Thus, multiple genetically defined subtypes of B-ALL are dependent on BET bromodomain activity in vitro.
Inhibition of proliferation and induction of apoptosis by JQ1 in B-ALL. (A) Dose-response of B-ALL cell line viability using a luminescent ATP detection assay with JQ1 treatment. Error bars represent SEM; n = 4. Inset: The chemical structure of JQ1. Table shows 50% growth inhibition values (GI50) and maximum effected cells (Emax) with annotated characteristic translocations for each cell line. (B) MHH-CALL4 and MUTZ-5 cell numbers with 500nM JQ1 treatment, normalized to time = 0 levels. Error bars represent SEM; n = 4 counts. (C) Caspase-3 and -7 activity and cell viability with 500 nm JQ1 treatment; data shown relative to vehicle control values and normalized to baseline time = 0 levels. Error bars represent SEM; n = 3 measurements. (D) Cellular apoptosis after treatment with DMSO (Veh) or 500nM JQ1 for 48 hours by flow cytometry with PI and AV. (E) PARP immunoblotting with 500nM JQ1 treatment. *Cleaved PARP band; β-tubulin (TUB) shown as loading control. (F) Cell cycle analysis of total DNA content by PI staining with 500nM JQ1 treatment.
Inhibition of proliferation and induction of apoptosis by JQ1 in B-ALL. (A) Dose-response of B-ALL cell line viability using a luminescent ATP detection assay with JQ1 treatment. Error bars represent SEM; n = 4. Inset: The chemical structure of JQ1. Table shows 50% growth inhibition values (GI50) and maximum effected cells (Emax) with annotated characteristic translocations for each cell line. (B) MHH-CALL4 and MUTZ-5 cell numbers with 500nM JQ1 treatment, normalized to time = 0 levels. Error bars represent SEM; n = 4 counts. (C) Caspase-3 and -7 activity and cell viability with 500 nm JQ1 treatment; data shown relative to vehicle control values and normalized to baseline time = 0 levels. Error bars represent SEM; n = 3 measurements. (D) Cellular apoptosis after treatment with DMSO (Veh) or 500nM JQ1 for 48 hours by flow cytometry with PI and AV. (E) PARP immunoblotting with 500nM JQ1 treatment. *Cleaved PARP band; β-tubulin (TUB) shown as loading control. (F) Cell cycle analysis of total DNA content by PI staining with 500nM JQ1 treatment.
Among the most sensitive B-ALL lines were MHH-CALL4 and MUTZ-5, which both harbor IGH-CRLF2 rearrangements.3 The CRLF2 locus is rearranged in 5%-10% of B-ALL through either IGH-CRLF2 translocation or an intrachromosomal P2RY8-CRLF2 deletion. Both rearrangements result in the overexpression of full-length CRLF2, which may confer a high risk of relapse.4-6,31-33 Because of the poor outcome among CRLF2-rearranged cases, we focused on this subtype to further explore mechanisms underlying JQ1 activity.
First, we determined the antiproliferative effects of JQ1 by counting treated cells in culture over the course of 10-day exposure (Figure 1B). Treatment with 500nM JQ1 reduced the proliferation of MHH-CALL4 and MUTZ-5 cells within 3 days and 3-5 days, respectively. We used 500nM JQ1, and this dose was used for similar assays in previous publications10,14 and thus allows for comparisons across malignant cell lineages.
Samples were taken during the time-course study to assay for the induction of apoptosis (by measuring caspase-3 and caspase-7 activity) as well as overall viability (by measuring cellular ATP levels; Figure 1C). Caspase activity in MHH-CALL4 cells was increased relative to vehicle-treated control cells after 24-hour exposure to JQ1 and by 3 days in MUTZ-5 cells. Viability in both cell lines was decreased within 24 hours of treatment and steadily declined over the subsequent 5 days. Caspase activity peaked after 3 days of exposure to JQ1 in MHH-CALL4 cells and after 5 days in MUTZ-5 cells, which corresponded to the initial decline of cell number in each line.
Next, we used flow cytometry after staining with propidium iodide (PI) and annexin V (AV) to assess the induction of cell death by JQ1 in MUTZ-5 and MHH-CALL4 cells. Treatment of both cell lines with 500nM JQ1 for 48 hours induced a significant decrease in viable (PIlow/AVlow) cells (P < .01), and an increase in apoptotic (PIhigh/AVhigh) cells (P < .01; Figure 1D; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Western blotting for PARP showed cleaved PARP, a marker of caspase-3 and -7 activity, appearing within 24 hours of JQ1 treatment in MHH-CALL4 cells and within 3 days in MUTZ-5 cells (Figure 1E). A time-course analysis of cell cycle after treatment with 500nM JQ1 showed no apparent effects on cell cycle progression after 24 hours in either cell line but an increased G1 fraction after 3 and 5 days of treatment (Figure 1F). To assess whether autophagy is a major component of JQ1-induced cell death in these lines, we performed immunoblotting for the autophagosome marker LC3 and found no effect on abundance or lipidation after 4 days of JQ1 treatment (supplemental Figure 2). Thus, apoptosis is the primary mechanism underlying the reduced viability of B-ALL cells exposed to JQ1, with a small but measurable G1 phase arrest that follows the induction of apoptosis.
JQ1 treatment decreases c-Myc and c-Myc target gene expression
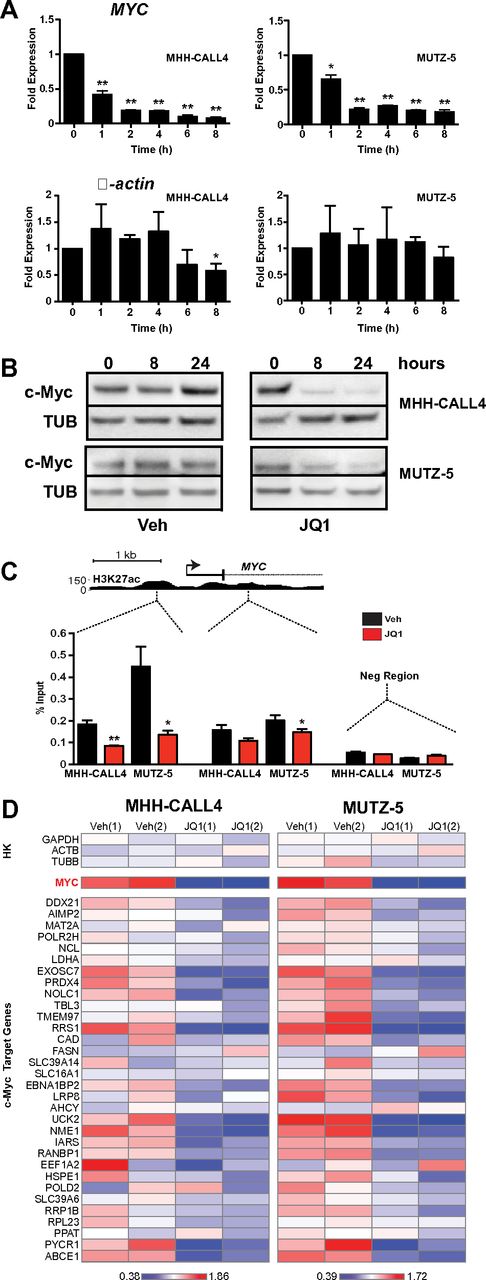
Recent studies of BET bromodomain inhibition identified a specific decrease in Myc transcription coupled with depletion of BRD4 at the MYC promoter.9-12 These observations support a model whereby BRD4 acts as a transcriptional coactivator that drives MYC expression. c-Myc is an oncogenic transcription factor that controls a gene expression program mediating cellular proliferation, metabolic adaptation, and survival.34 Thus, we hypothesized that JQ1 treatment would decrease c-Myc expression and thereby suppress transcription of c-Myc–dependent target genes in B-ALL. Treatment of both IGH-CRLF2–rearranged cell lines with 500nM JQ1 induced rapid and sustained decreases in MYC transcript levels (Figure 2A). This was accompanied by a loss of detectable c-Myc protein in whole cell lysates at both 8 and 24 hours after treatment (Figure 2B). ChIP using an anti-BRD4 antibody followed by quantitative PCR demonstrated BRD4 localization at MYC promoter-enhancer elements in both lines and depletion with exposure to JQ1 (Figure 2C; replicate experiment shown in supplemental Figure 3).
JQ1 decreases c-Myc expression in CRLF2-rearranged B-ALL. (A) Quantitative PCR analysis of MYC transcript levels in MHH-CALL4 and MUTZ-5 B-ALL cells treated with 500nM JQ1. β-actin expression shown as control. Data for each time point are normalized to vehicle control and presented as the ratio of expression compared with baseline expression at time = 0. Error bars represent ± SEM. *P < .1 (paired t test). **P < .01 (paired t test). (B) Immunoblotting for c-Myc in whole cell lysates of cells treated with 500nM JQ1. (C) ChIP with a BRD4 antibody at 2 sites within the MYC promoter region in cells treated with 500nM JQ1 for 4 hours. Enrichment is shown as the percentage of total input DNA. The negative control region primers amplify within a gene desert ∼ 1 Mb upstream of MYC. (D) Heatmaps of MYC and c-Myc target gene expression in MHH-CALL4 and MUTZ-5 cells treated with 500nM JQ1 for 4 hours. Scale bars represent fold expression changes of each gene relative to the average gene expression across the 4 samples. HK indicates housekeeping controls. Data for 2 biologic replicates are shown.1,2
JQ1 decreases c-Myc expression in CRLF2-rearranged B-ALL. (A) Quantitative PCR analysis of MYC transcript levels in MHH-CALL4 and MUTZ-5 B-ALL cells treated with 500nM JQ1. β-actin expression shown as control. Data for each time point are normalized to vehicle control and presented as the ratio of expression compared with baseline expression at time = 0. Error bars represent ± SEM. *P < .1 (paired t test). **P < .01 (paired t test). (B) Immunoblotting for c-Myc in whole cell lysates of cells treated with 500nM JQ1. (C) ChIP with a BRD4 antibody at 2 sites within the MYC promoter region in cells treated with 500nM JQ1 for 4 hours. Enrichment is shown as the percentage of total input DNA. The negative control region primers amplify within a gene desert ∼ 1 Mb upstream of MYC. (D) Heatmaps of MYC and c-Myc target gene expression in MHH-CALL4 and MUTZ-5 cells treated with 500nM JQ1 for 4 hours. Scale bars represent fold expression changes of each gene relative to the average gene expression across the 4 samples. HK indicates housekeeping controls. Data for 2 biologic replicates are shown.1,2
To define the effects of JQ1 treatment on the transcription of c-Myc target genes, we developed a multiplexed transcript detection assay that includes 32 known c-Myc target genes. Treatment with 500nM JQ1 for 4 hours resulted in MYC down-regulation as well as a significant decrease in expression of the c-Myc target gene set (P < .01 by 2-way ANOVA) in both MHH-CALL4 and MUTZ-5 cells (Figure 2D). Importantly, the expression of housekeeping gene controls was relatively unchanged, highlighting the specific transcriptional effects of BET bromodomain inhibition. P values of expression changes for each individual gene are shown in supplemental Figure 4A. Replicate data from 2 separate experiments were well correlated (R2 > 0.97; supplemental Figure 4B), and unsupervised hierarchical clustering of the c-Myc target gene dataset segregated each replicate experiment correctly by cell line (supplemental Figure 5). Among the c-Myc targets with greatest relative down-regulation on treatment with JQ1 were NME1, a leukemia cell differentiation inhibitor and marker of poor prognosis in ALL,35,36 EXOSC7, a component of an antiapoptotic transcriptional repressor complex,37 the uridine/cytidine kinase UCK2,38 2 genes involved in ribosome biogenesis and perhaps telomere clustering (RRS1 and EBNA1BP2),39 and the proline synthesis pathway component PYCR1.40
JQ1 treatment decreases IL7R expression and JAK2/STAT5 activation
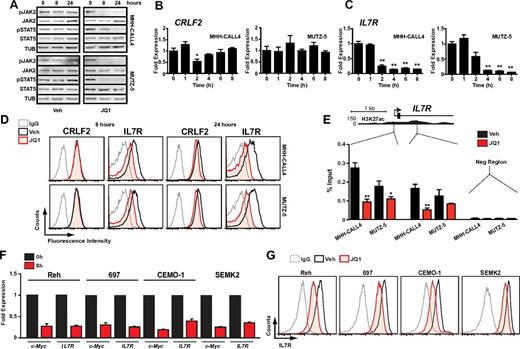
MUTZ-5 and MHH-CALL4 were among the most sensitive B-ALL cell lines to JQ1 (Figure 1A), suggesting that they either have particular dependence on c-Myc or that JQ1 targets factors of selective importance within these lines. In nonmalignant hematopoietic cells, CRLF2 forms heterodimers with the IL7R and signals in response to thymic stromal lymphopoietin (TSLP) through activation of JAK/STAT signaling. In B-ALL cells with CRLF2 rearrangements, the specific role of CRLF2/IL7R heterodimers has not been resolved, as these leukemias commonly harbor activating mutations in CRLF2, JAK2, JAK1, or rarely IL7R that may obviate the need for the full receptor complex.3,5,41,42 Indeed, both MHH-CALL4 and MUTZ-5 harbor JAK2 exon 16 mutations in addition to IGH-CRLF2 translocations.3 To determine whether JQ1 treatment affects CRLF2/IL7R signaling, we treated both lines with JQ1 and assayed phosphorylation at activation sites in JAK2 and STAT5. As noted in Figure 3A, treatment with 500nM JQ1 almost completely eliminated phosphorylation of JAK2 and STAT5 after 8 and 24 hours.
Bromodomain inhibition decreases IL7R expression and suppresses JAK2/STAT5 phosphorylation. (A) Immunoblotting of whole cell lysates from cells treated with 500nM JQ1 at indicated time points. (B) Quantitative PCR of CRLF2 transcript levels in MHH-CALL4 and MUTZ-5 cells treated with 500nM JQ1. Data for each time point are normalized to vehicle control and presented as a ratio of expression compared with baseline expression at time = 0. Error bars represent ± SEM. *P < .1 (paired t test). **P < .01 (paired t test). (C) Quantitative PCR analysis of IL7R transcripts levels, as in panel B. (D) Flow cytometry for CRLF2 and IL7R. Cells were treated with 500nM JQ1 or vehicle for 8 and 24 hours and stained with an antibody against the indicated target or an isotype IgG. (E) ChIP with a BRD4 antibody at 2 sites within the IL7R promoter region in cells treated with 500nM JQ1 for 4 hours. Enrichment is shown as the percentage of total input DNA. Negative control region primers amplify within a gene desert region ∼ 70 kb downstream of IL7R. *P < .1 (unpaired t test). **P < .01 (unpaired t test). (F) Quantitative PCR of MYC and IL7R expression and (G) flow cytometry of IL7R expression in B-ALL cell lines that lack CRLF2 rearrangements after 8-hour treatment with 500nM JQ1 or vehicle, as above.
Bromodomain inhibition decreases IL7R expression and suppresses JAK2/STAT5 phosphorylation. (A) Immunoblotting of whole cell lysates from cells treated with 500nM JQ1 at indicated time points. (B) Quantitative PCR of CRLF2 transcript levels in MHH-CALL4 and MUTZ-5 cells treated with 500nM JQ1. Data for each time point are normalized to vehicle control and presented as a ratio of expression compared with baseline expression at time = 0. Error bars represent ± SEM. *P < .1 (paired t test). **P < .01 (paired t test). (C) Quantitative PCR analysis of IL7R transcripts levels, as in panel B. (D) Flow cytometry for CRLF2 and IL7R. Cells were treated with 500nM JQ1 or vehicle for 8 and 24 hours and stained with an antibody against the indicated target or an isotype IgG. (E) ChIP with a BRD4 antibody at 2 sites within the IL7R promoter region in cells treated with 500nM JQ1 for 4 hours. Enrichment is shown as the percentage of total input DNA. Negative control region primers amplify within a gene desert region ∼ 70 kb downstream of IL7R. *P < .1 (unpaired t test). **P < .01 (unpaired t test). (F) Quantitative PCR of MYC and IL7R expression and (G) flow cytometry of IL7R expression in B-ALL cell lines that lack CRLF2 rearrangements after 8-hour treatment with 500nM JQ1 or vehicle, as above.
In our previous study of multiple myeloma, JQ1 treatment decreased c-Myc transcription in lines that harbor an IGH-MYC translocation10 concomitant with a decrease in BRD4 binding to both the MYC promoter and a transcriptional enhancer region within the IGH locus. Thus, we hypothesized that JQ1 treatment decreases CRLF2 expression through its association with the IGH locus in MHH-CALL4 and MUTZ-5 cells. However, treatment of each line with 500nM JQ1 over 8 hours did not affect CRLF2 expression (Figure 3B). In contrast, IL7R transcript levels decreased markedly in both lines on treatment with JQ1 (Figure 3C). Flow cytometry revealed a 60%-70% decrease in surface IL7R after 24 hours of JQ1 treatment, with a much smaller reduction in surface CRLF2 (Figure 3D; replicate experiment and quantification shown in supplemental Figure 6). Analysis of BRD4 binding to the IL7R locus revealed its localization to active cis elements within the IL7R promoter region by ChIP (Figure 3E; supplemental Figure 7). BRD4 was effectively depleted from the IL7R promoter region on exposure to 500nM JQ1 (Figure 3E; supplemental Figure 7), the same dose that promoted down-regulation of IL7R protein (Figure 3D).
In the recently published Cancer Cell Line Encyclopedia,43 B-ALL cell lines had the highest mean expression of IL7R compared with other cancer types (supplemental Figure 8). Thus, we investigated whether BET inhibition could affect IL7R expression in other B-ALL subtypes or whether this is specific to CRLF2-rearranged cells. We treated 4 additional B-ALL cell lines with 500nM JQ1. After 8 hours, substantial suppression of both MYC and IL7R transcripts was observed in all 4 lines (Figure 3F), with similar decreases in surface IL7R expression (Figure 3G). Together, these data demonstrate that JQ1 suppresses a major B-ALL oncogenic signaling pathway involving JAK2-STAT5 in CRLF2-rearranged leukemia cells, at least in part by targeting the transcriptional activation of IL7R. Furthermore, JQ1 can broadly suppress IL7R and MYC transcription in B-ALL.
Analysis of differential gene expression with short-term JQ1 treatment
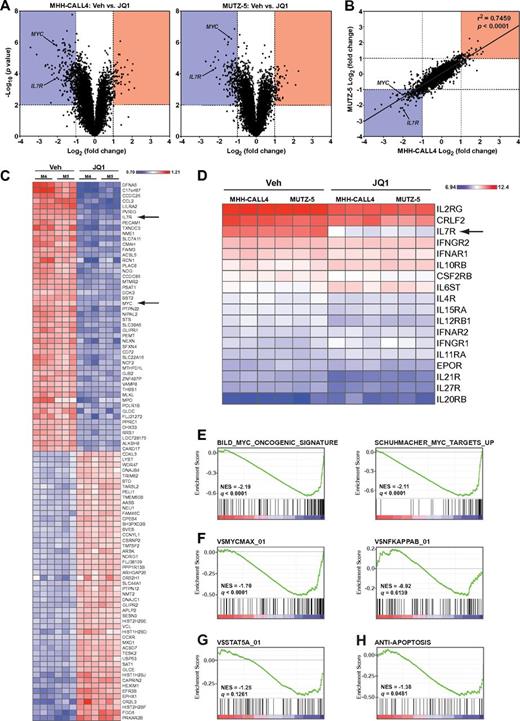
To determine the early transcriptional consequences of BET inhibition, we performed gene expression profiling of MUTZ-5 and MHH-CALL4 cells 8 hours after exposure to JQ1 or vehicle. In both cell lines, JQ1 treatment induced expression changes in a relatively small subset of genes, as opposed to a general disruption of global transcription (Figure 4A). Expression changes in both cell lines were well correlated, with MYC and IL7R being among the most statistically significant down-regulated genes in both lines (Figure 4A-B). When both lines were analyzed together, IL7R and MYC were the seventh and 23rd most suppressed genes, respectively, on exposure to JQ1 (Figure 4C). Strikingly, IL7R was the only type I or type II cytokine receptor with significantly reduced expression on treatment with JQ1 (Figure 4D). Although both IL7R heterodimerization partners (IL2RG and CRLF2) are highly expressed in MHH-CALL4 and MUTZ-5 cells, their transcript levels were not significantly reduced by exposure to JQ1 (Figure 4D).
Genome-wide transcriptome analysis of JQ1-treated cells. (A) Volcano plots of gene expression differences for MHH-CALL4 and MUTZ-5 cells treated with vehicle or 500nM JQ1 for 8 hours. (B) Correlation of gene expression changes between MHH-CALL4 and MUTZ-5 cells treated with JQ1. (C) Heatmap of the top 50 significantly up- and down-regulated genes. P < .01. Each gene row is normalized to mean expression level (M4 = MHH-CALL4; M5 = MUTZ-5). (D) RMA-normalized expression values for all expressed cytokine receptors. (E) Gene set enrichment analysis of 2 c-Myc signatures. (F) c-Myc and NF-κB binding site gene sets. (G) STAT5A binding site gene set. (H) An antiapoptosis gene set. NES indicates normalized enrichment score; and q = false discovery rate.27
Genome-wide transcriptome analysis of JQ1-treated cells. (A) Volcano plots of gene expression differences for MHH-CALL4 and MUTZ-5 cells treated with vehicle or 500nM JQ1 for 8 hours. (B) Correlation of gene expression changes between MHH-CALL4 and MUTZ-5 cells treated with JQ1. (C) Heatmap of the top 50 significantly up- and down-regulated genes. P < .01. Each gene row is normalized to mean expression level (M4 = MHH-CALL4; M5 = MUTZ-5). (D) RMA-normalized expression values for all expressed cytokine receptors. (E) Gene set enrichment analysis of 2 c-Myc signatures. (F) c-Myc and NF-κB binding site gene sets. (G) STAT5A binding site gene set. (H) An antiapoptosis gene set. NES indicates normalized enrichment score; and q = false discovery rate.27
To assess the influence of BET inhibition on specific transcriptional pathways, we evaluated 2 established MYC gene expression signatures44,45 and found a significant negative enrichment for both in cells treated with JQ1 (Figure 4E). Moreover, JQ1 treatment significantly down-regulated genes containing MYC binding sites (V$MYCMAX_01) but did not significantly affect the NF-κB pathway (V$NFKAPPAB_01; Figure 4F). A set of genes containing STAT5A binding sites (V$STAT5A_01) was also down-regulated in cells treated with JQ1 (Figure 4G). Lastly, given our observation that apoptosis is a primary effect of BET inhibition in these cells, we examined a signature of antiapoptotic genes (ANTI_APOPTOSIS) annotated by the Gene Ontology project (GO term 0006916). This signature was also significantly enriched, with down-regulation of genes, such as BCL2, that were previously shown to be affected by BET inhibition (Figure 4H).12
Antileukemic effects of JQ1 in an in vivo model of primary CRLF2-rearranged B-ALL
To assess the in vivo potential of BET bromodomain inhibition, we used a murine xenograft model of primary CRLF2-rearranged B-ALL.26 NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice were engrafted with a primary B-ALL sample that harbors a P2RY8-CRLF2 rearrangement and lacks somatic mutation within the known components of CRLF2 signaling, including IL7R, CRLF2, TSLP, JAK1, JAK2, and STAT5A/B. Moreover, this sample lacks kinase fusions that could activate JAK signaling (eg, BCR-JAK2) and has a normal karyotype.26 Secondary transplantation was performed to expand cohorts for study, and sentinel mice were killed after 4 weeks to confirm > 30% bone marrow involvement by human CD45+ cells (supplemental Figure 9). At that point, mice were randomized to receive once-daily intraperitoneal injections with either JQ1 (50 mg/kg) or vehicle.
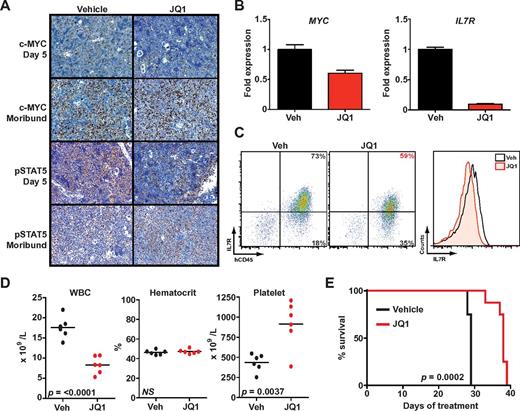
After 5 days of treatment with JQ1, spleens from killed animals exhibited marked reductions in c-Myc and phospho-STAT5–expressing cells compared with vehicle-treated mice using established IHC protocols (Figure 5A).29,30 RT-PCR analysis of MYC and IL7R expression in leukemia cells harvested from spleens revealed significant decreases in both transcripts (Figure 5B). Furthermore, surface IL7R expression was reduced on human CD45+ leukemia cells from mice treated with JQ1 compared with mice treated with vehicle (Figure 5C). In contrast, CRLF2 expression did not differ between treatment arms, which accords with the observations seen in our cell line experiments (supplemental Figure 10). After 4 weeks of treatment, mice receiving JQ1 had lower white blood cell and higher platelet counts compared with vehicle-treated mice (Figure 5D), consistent with reduced leukemic burden in the peripheral blood and bone marrow. Genetic studies of BRD3 have suggested a mechanistic role in erythropoiesis,46 yet mice who received BET bromodomain inhibition for 4 weeks did not have significant anemia (Figure 5D).
JQ1 decreases leukemic burden and improves survival in B-ALL primary human xenografts. (A) Immunohistochemistry for c-Myc30 and phospho-STAT5 (pSTAT5) in spleen samples obtained after 5-day treatment or at the moribund state. (B) Quantitative PCR of MYC and IL7R expression in leukemia cells harvested from spleens of mice treated for 5 days with JQ1 or vehicle (Veh). Error bars represent ± SEM; n = 3 PCR samples normalized to vehicle-treated sample. (C) Flow cytometry of splenic cells obtained after 5-day treatment with JQ1 or vehicle. Scatter plots on the left show overall IL7R expression versus hCD45. The histogram on the right is a quantification of total IL7R expression. (D) Peripheral blood cell counts after 5 days of treatment (n = 6 in each arm, mean levels for each sample marked). WBC indicates white blood cells. (E) Survival of engrafted mice treated with JQ1 (n = 9) or vehicle (n = 9).
JQ1 decreases leukemic burden and improves survival in B-ALL primary human xenografts. (A) Immunohistochemistry for c-Myc30 and phospho-STAT5 (pSTAT5) in spleen samples obtained after 5-day treatment or at the moribund state. (B) Quantitative PCR of MYC and IL7R expression in leukemia cells harvested from spleens of mice treated for 5 days with JQ1 or vehicle (Veh). Error bars represent ± SEM; n = 3 PCR samples normalized to vehicle-treated sample. (C) Flow cytometry of splenic cells obtained after 5-day treatment with JQ1 or vehicle. Scatter plots on the left show overall IL7R expression versus hCD45. The histogram on the right is a quantification of total IL7R expression. (D) Peripheral blood cell counts after 5 days of treatment (n = 6 in each arm, mean levels for each sample marked). WBC indicates white blood cells. (E) Survival of engrafted mice treated with JQ1 (n = 9) or vehicle (n = 9).
Treatment with JQ1 significantly prolonged overall survival compared with vehicle (P = .0002, Figure 5E). Immunohistochemistry of moribund mice in both arms demonstrated extensive involvement of spleens with leukemia that highly expressed both c-Myc and phospho-STAT5. Thus, failure among JQ1-treated mice is associated with reactivation of the same signaling pathways. To rule out mutations in BRD4 as a mechanism of resistance, we sequenced exons 2, 3, 4, and 6 of BRD4, which contain the bromodomain, from leukemia cells of moribund mice treated with JQ1. No somatic base changes were detected.
Discussion
Oncogenic dysregulation of cellular growth pathways, mediated and reinforced by chromatin modification and associated factors, is essentially ubiquitous across cancer subtypes.47 This understanding has been accompanied by the emergence of a limited number of promising therapeutic agents specifically designed to target aberrant gene regulatory factors.48 These factors generally modify either chromatin or DNA by adding (“writing”) or removing (“erasing”) specific chemical moieties involved in oncogenic gene expression programs. The recent discovery that bromodomains, the recognition modules or “readers” of acetyllysines, are protein targets amenable to small-molecule inhibition has led to their investigation as cancer dependencies.9-12,14
Here we report data supporting bromodomain inhibition as a novel therapeutic strategy for B-ALL. Using JQ1, a first-in-class bromodomain prototype drug developed in the Bradner laboratory, we demonstrate potent activity against a range of human B-ALL cell lines. Significantly, BET bromodomain inhibition was effective at reducing the viability of B-ALL cells with several different oncogenic “drivers,” including the only known CRLF2-rearranged cell lines, chemotherapy-refractory ETV6-PDGFRB–rearranged NALM-6 cells, CEMO-1 cells that harbor an IGH-BCL9 rearrangement, the relapsed B-ALL line 697 with a TCF3-PBX1 rearrangement, and 2 MLL-AF4–harboring lines.
We also show, for the first time, that JQ1 has in vivo activity against primary human leukemia. However, all mice treated with JQ1 ultimately progressed and died of xenografted leukemia, at which time both c-Myc and p-STAT5 were abundantly reactivated. Progression did not result from the acquisition of resistance mutations within the BRD4 bromodomain, suggesting that alternate mechanisms underlie the progression of disease. These data then prompt the study and development of drug-like derivatives of JQ1 with improved pharmacologic properties while exploring in detail mechanisms of resistance such as overexpression of drug efflux pumps and activation of orthogonal growth pathways.
Despite previous evidence that BRD4 inhibition can reduce the expression of oncogenes translocated to the IGH locus,10,11 CRLF2 gene expression was not significantly reduced on JQ1 treatment of either MUTZ5 or MHH-CALL4 cells. The difference in effects on IGH-driven oncogene expression could simply reflect heterogeneity between cell lines or it could indicate that BRD4 involvement in IGH transcription may differ based on the stage of B-cell ontogeny. Whereas IGH-MYC translocations occur within mature B cells undergoing class switch recombination, IGH-CRLF2 translocations (as well as other IGH rearrangements in B-ALL) occur within pre/pro-B cells undergoing V(D)J recombination.49
A novel finding in this study is the down-regulation of IL7R transcription by BET bromodomain inhibition. IL7R expression is restricted to the lymphoid lineage and is required for normal lymphoid development.50 In normal hematopoietic cells, IL7R heterodimerizes with CRLF2 to form a receptor for TSLP and also heterodimerizes with IL2RG (γc) to form a receptor for IL7. However, the precise relationship between IL7R expression and CRLF2 activity in the context of CRLF2-rearranged B-ALL has not been conclusively determined. Whereas extensive studies will be necessary to understand the action of these cytokine receptors in B-ALL, several lines of evidence show that IL7R can play a substantial role in oncogenic JAK/STAT signaling. First, CRLF2 does not undergo measurable homodimerization, except in the context of an F232C mutation, which generates an intermolecular disulfide bond.5 In addition, exposure of Ba/F3 cells that markedly overexpress wild-type CRLF2 to TSLP does not appreciably support cytokine-independent viability or phosphorylation of JAK2 and STAT5.2,5 Thus, CRLF2 alone cannot significantly promote signaling, either in the presence or absence of TSLP. Strikingly, our gene expression analysis in this study did not reveal expression changes in either CRLF2, IL2RG, or any other type I or type II cytokine receptor on exposure to JQ1. Together, this experimental evidence indicates a requirement for IL7R to support JAK2/STAT5 signaling in these cells. MUTZ-5 and MHH-CALL4 cells are exceptionally refractory to lipid-based transfection techniques and are not amenable to antibiotic-based selection after viral transduction. To date, our attempts to knock down IL7R using both siRNA and shRNA in MUTZ-5 and MHH-CALL4 cells have been technically unsuccessful.
Activating mutations in IL7R were recently identified in 6% of B-ALL cases with overexpression of CRLF2 and at least 9% of pediatric T-lineage ALL cases.41,51 Furthermore, dysregulated IL7R activity plays a role in some autoimmune disorders.52 Thus, BET bromodomain inhibition may be an effective strategy in CRLF2-rearranged B-ALL, some cases of T-lineage ALL, as well as other disorders dependent on IL7R.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank R. Zeid for assistance with the multiplexed gene expression experiments, P. Rahl for help curating the c-Myc target gene set, and A. Christie, A. Lane, and D. van Bodegom for assistance with mouse experiments.
This work was supported by the National Institutes of Health/National Cancer Institute (R01 CA151898-01, D.M.W.), the Leukemia & Lymphoma Society (C.J.O. and J.E.B.), a Damon-Runyon Cancer Research Foundation Innovator Award (J.E.B.), and the American Society of Hematology (Junior Faculty Scholar Awards, J.E.B. and D.M.W.).
National Institutes of Health
Authorship
Contribution: C.J.O. and N.K. designed and performed research, analyzed data, and wrote the paper; L.B. and R.M.P. designed and performed research; J.Q. contributed vital new reagents; T.B. performed research; S.J.R. performed contributed vital new reagents and analyzed data; A.L.K. performed research and analyzed data; J.E.B. designed research, contributed vital new reagents, and analyzed data; and D.M.W. designed research, contributed vital new reagents, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: J.E.B. and the Dana-Farber Cancer Institute have founded Tensha Therapeutics to translate drug-like inhibitors of BET bromodomains as cancer therapeutic agents. The remaining authors declare no competing financial interests.
Correspondence: David Weinstock, Dana-Farber Cancer Institute, 450 Brookline Ave, Dana 510B, Boston, MA 02215; e-mail: dweinstock@partners.org; and James E. Bradner, Dana-Farber Cancer Institute, 450 Brookline Ave, Dana 510D, Boston, MA 02215; e-mail: james_bradner@dfci.harvard.edu.
References
Author notes
C.J.O. and N.K. contributed equally to this study.
J.E.B. and D.M.W. contributed equally to this study.