Key Points
ERG overexpression in transgenic mice induces a transcriptional leukemia stem cell program characteristic of human AML.
PIM1 and RAS are relevant ERG therapeutic targets.
Abstract
The ETS transcription factor ERG plays a central role in definitive hematopoiesis, and its overexpression in acute myeloid leukemia (AML) is associated with a stem cell signature and poor prognosis. Yet how ERG causes leukemia is unclear. Here we show that pan-hematopoietic ERG expression induces an early progenitor myeloid leukemia in transgenic mice. Integrated genome-scale analysis of gene expression and ERG binding profiles revealed that ERG activates a transcriptional program similar to human AML stem/progenitor cells and to human AML with high ERG expression. This transcriptional program was associated with activation of RAS that was required for leukemia cells growth in vitro and in vivo. We further show that ERG induces expression of the Pim1 kinase oncogene through a novel hematopoietic enhancer validated in transgenic mice and human CD34+ normal and leukemic cells. Pim1 inhibition disrupts growth and induces apoptosis of ERG-expressing leukemic cells. The importance of the ERG/PIM1 axis is further underscored by the poorer prognosis of AML highly expressing ERG and PIM1. Thus, integrative genomic analysis demonstrates that ERG causes myeloid progenitor leukemia characterized by an induction of leukemia stem cell transcriptional programs. Pim1 and the RAS pathway are potential therapeutic targets of these high-risk leukemias.
Introduction
The ETS transcription factor ERG has been implicated as a major regulator of both normal and aberrant hematopoiesis.1,2 Further insights into ERG function in normal hematopoiesis have come from genome-wide binding site analysis, which revealed that ERG takes part in a heptad of transcription factors that preferentially bind to hematopoietic enhancers in the mouse multipotent hematopoietic progenitor cell line HPC-7.3
Aberrant ERG expression is strongly linked to cancer, as highlighted by its frequent involvement in chromosomal translocations associated with various malignancies such as TMPRSS2-ERG in prostate cancer, EWS-ERG in sarcoma, and TLS-ERG in leukemia.4-6 We have previously shown that ERG is a megakaryocytic oncogene.7 Moreover, ERG serves as an independent prognostic factor in cytogenetically normal acute myeloid leukemia (AML), and its expression is positively correlated with adverse outcome in both T-cell acute lymphocytic leukemia (T-ALL) and AML.8-10 ERG is also included in a human leukemia stem cell gene signature that correlates with a worse outcome in AML patients.11 Despite this substantial evidence implicating ERG in leukemia development and maintenance, little is known about the molecular mechanisms used by ERG in leukemic cells.
To address this issue, we generated transgenic mice with pan-hematopoietic ERG expression. Similar to human leukemias with increased expression of ERG, TgERG mice develop either T-lymphoid12 or myeloid acute leukemias by 5 months of age. Through combined gene expression and chromatin immunoprecipitation-sequencing (ChIP-Seq) profiling, we now show that ERG overexpression in myeloid leukemias activates a stem cell signature characteristic of human AMLs. We also identify the oncogenic PIM1 kinase as a direct ERG target through its binding to a novel enhancer, and the RAS pathway as an indirect target of ERG. Finally, we demonstrate that pharmacologic inhibition of either of these targets is therapeutically relevant.
Methods
Transgenic mice and xenografts
TgERG mice were generated as previously described.12 For transplantations, single cells were prepared from the spleens of TgERG leukemic mice, washed in phosphate-buffered saline, and injected (5*105 per mouse) into the tail veins of NOD scid Il2rgnull (NSG) mice.
Histology
Spleen and liver tissues were fixed in 4% neutral buffered formalin, transferred to 70% ethanol the next day, paraffin-embedded, and stained with hematoxylin and eosin using the standard protocols. Bone marrow cells were cytocentrifuged, fixed, and stained with May Grunwald/Giemsa stain (Sigma-Aldrich).
Immunophenotyping
Leukemic blasts taken from the bone marrow of TgERG mice were washed in phosphate-buffered saline with 0.05M ethylenediamine tetraacetic acid and 0.1% bovine serum antigen and then lineage depleted using a lineage depletion kit (Miltenyi Biotec). Lineage-depleted leukemia cells were stained with PE-Cy7–conjugated anti–c-Kit, FITC-conjugated anti-Sca1, and APC-conjugated anti-CD150 antibodies (eBioscience). Cells were subsequently analyzed using the Gallios Flow cytometer and Kaluza Flow Analysis Software (Beckman Coulter, Inc.).
Gene expression profiling
Experiments were performed using Affymetrix Mouse gene 1.0 ST oligonucleotide arrays (Affymetrix, Santa Clara, CA). RNA samples were prepared from the bone marrow of 3 TgERG mice (generated from 2 different founder lines), 3 wild-type (WT) littermates, and 3 pools of lineage-depleted WT bone marrow cells. Total RNA from each sample was used to prepare biotinylated target cDNA according to the manufacturer’s recommendations. A detailed description of the method is available in the supplemental Methods. Raw data have been submitted to the National Center for Biotechnology Information to be accessed via the Gene Expression Omnibus portal (www.ncbi.nlm.nih.gov/geo; GEO record GSE49787)
Chromatin immunoprecipitation
ChIP material was prepared from the spleens of 2 TgERG mice with AML of identical immunophenotype; Tg1 on the background of WT Gata112 and Tg 2 on Gata1s background.13 ChIP assays were performed as previously described using a rabbit polyclonal antibody raised against Erg-1/2/3 (clone C-17, Santa Cruz). As a control, nonspecific rabbit IgG (I5006; Sigma Aldrich) was used. Enrichment was first validated by reverse-transcriptase polymerase chain reaction (RT-PCR) with the following primers: Hhex+1 forward; 5′-CGACGTCTGATAGCCAGGAT-3′, reverse; 5′-GAGAGGCAGAGAGGAAGCAA-3′. ChIP samples were amplified and sequenced using the Illumina GII Genome Analyzer, following the manufacturer’s instructions. Raw and mapped sequence data have been submitted to the National Center for Biotechnology Information to be accessed via the Gene Expression Omnibus portal (www.ncbi.nlm.nih.gov/geo; GEO record GSE46554). Additional details can be found in the supplemental Methods.
Enhancer activity assays
The Pim1 +10-kb region was amplified from mouse genomic DNA using the restriction site introducing primers: cgcggatccGGGTTCAGATTAGCCATACCC-acgcgtcgacAACACCCTACCCAGGGCTTC. This fragment was inserted downstream of the luciferase gene in pGL2 (Promega) or downstream from an SV40 minimal promoter in a construct containing a lacZ reporter cassette. The mutation constructs were made as previously described14 and verified by sequencing. ETS site mutations were generated by PCR and verified by sequencing.
L8057 cells15 were grown in Dulbecco’s modified Eagle’s medium/F-12 with glutamax (Invitrogen) in 15% fetal calf serum. Stable transfections and luciferase assays were performed as described.16
F0 transgenic embryos were generated by pronuclear injection of reporter constructs by Cyagen Biosciences Inc. (Guangzhou, China). Embryos were harvested at embryonic days 11 to 12.5 and analyzed as described.17 All animal studies were performed according to UK Home Office guidelines with Home Office approval.
Drugs
Farnesyl thiosalicylic acid (FTS) (Salirasib) was kindly provided by Yoel Kloog (Tel-Aviv University). SGI-1776 was purchased from Selleck chemicals. For in vitro assays, both compounds were dissolved in dimethyl sulfoxide, and final concentrations were adjusted to 0.1%. For in vivo assays, FTS was prepared as described.18
Knockdown experiments
Stable knockdown of ERG in CMY cells was performed as previously described.7 For PIM1 knockdown, TgERG leukemia cells (2.5*106) were transiently transfected with Pim1 siRNA oligonucleotides or with Cy-5-conjugated nontargeting siRNA oligonucleotides (Sigma Aldrich) using AMAXA Nucleofector Kit V (Lonza) according to the manufacturer’s instructions and by performing 2 consecutive pulses 24 hours apart.
Results
Transgenic ERG mice develop acute leukemia originating from an early myeloid progenitor
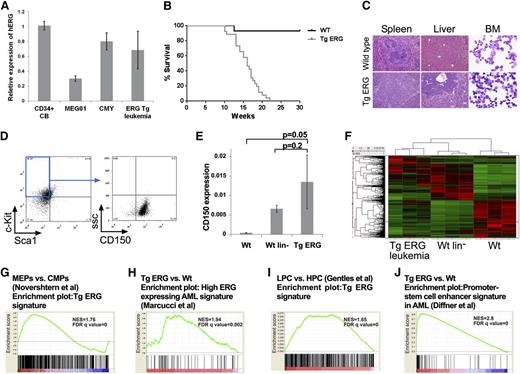
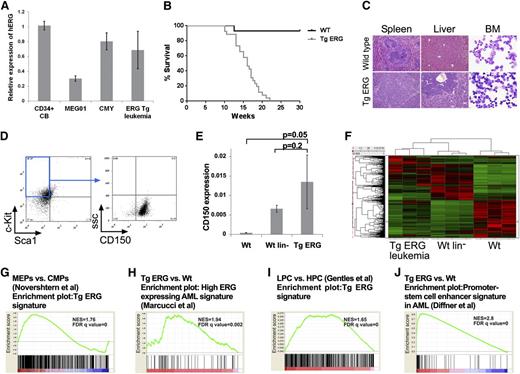
To examine the leukemogenic potential of ERG, we generated transgenic mice expressing the human ERG3 hematopoietic isoform under the control of the vav promoter. ERG expression levels in the transgenic leukemia are within the range found in normal human CD34+ hematopoietic progenitors or in human AML cells (Figure 1A). We previously reported that ∼30% of TgERG mice develop T-ALL.12 All other TgERG mice died from myeloid leukemia by the age of 5 months (Figure 1B), manifested by massive infiltration of leukemic blasts into the bone marrow, the spleen, and the liver (Figure 1C) and by marked anemia and thrombocytopenia (supplemental Figure 1). Immune phenotyping showed leukemic cells to be lineage negative and c-Kit+ Sca-1– CD150+, characteristic of megakaryocyte-erythroid progenitors (MEPs)22-24 (Figure 1D). Consistent with the progenitor origin of TgERG leukemias, CD150 expression was similar to lineage-negative (lin–) cells and significantly elevated compared with whole bone marrow (Figure 1E).
ERG induces an early progenitor myeloid leukemia in mice. (A) Bar graph depicting expression of human ERG by quantitative RT-PCR in TgERG leukemia and human AML cell lines MEG01 and CMY compared with normal CD34+ cells derived from cord blood (mean ± SEM, n = 3; experiment was repeated twice). (B) Kaplan-Meier survival curve showing that all transgenic ERG mice die by the age of 22 weeks (WT mice, n = 14; TgERG mice, n = 26). (C) Hematoxylin and eosin–stained sections of spleens and livers and Giemsa-stained cytospin samples from the bone marrow of TgERG mice and WT littermates (original magnification ×200 for sections, ×600 for cytospin samples). (D) Immunophenotype of TgERG leukemias. Typical dot plots showing that most of the bone marrow cells found in sick TgERG mice are lin–, cKit+ Sca-1– CD150+. (E) Validation of CD150 expression by quantitative RT-PCR normalized to β-actin (mean ± SEM, n = 3 for WT and WT lin–. N = 5 for TgERG leukemia). (F) Heat map depicting hierarchical clustering of TgERG leukemias and WT controls (see supplemental Methods). (G) GSEA showing enrichment of the TgERG leukemia signature in MEPs compared with CMPs.19 (H) GSEA showing enrichment of high ERG–expressing human AML signature9 in the TgERG leukemia compared with WT mice. (I) GSEA showing enrichment of TgERG leukemia signature in human AML progenitors compared with normal hematopoietic progenitors.20 (J) GSEA showing enrichment of the promoter-stem cell enhancer signature21 in TgERG leukemia compared with WT mice.
ERG induces an early progenitor myeloid leukemia in mice. (A) Bar graph depicting expression of human ERG by quantitative RT-PCR in TgERG leukemia and human AML cell lines MEG01 and CMY compared with normal CD34+ cells derived from cord blood (mean ± SEM, n = 3; experiment was repeated twice). (B) Kaplan-Meier survival curve showing that all transgenic ERG mice die by the age of 22 weeks (WT mice, n = 14; TgERG mice, n = 26). (C) Hematoxylin and eosin–stained sections of spleens and livers and Giemsa-stained cytospin samples from the bone marrow of TgERG mice and WT littermates (original magnification ×200 for sections, ×600 for cytospin samples). (D) Immunophenotype of TgERG leukemias. Typical dot plots showing that most of the bone marrow cells found in sick TgERG mice are lin–, cKit+ Sca-1– CD150+. (E) Validation of CD150 expression by quantitative RT-PCR normalized to β-actin (mean ± SEM, n = 3 for WT and WT lin–. N = 5 for TgERG leukemia). (F) Heat map depicting hierarchical clustering of TgERG leukemias and WT controls (see supplemental Methods). (G) GSEA showing enrichment of the TgERG leukemia signature in MEPs compared with CMPs.19 (H) GSEA showing enrichment of high ERG–expressing human AML signature9 in the TgERG leukemia compared with WT mice. (I) GSEA showing enrichment of TgERG leukemia signature in human AML progenitors compared with normal hematopoietic progenitors.20 (J) GSEA showing enrichment of the promoter-stem cell enhancer signature21 in TgERG leukemia compared with WT mice.
To further characterize TgERG myeloid leukemias, we performed gene expression profiling on 3 TgERG bone marrow leukemia samples, 3 WT littermates, and 3 pools of WT lin– cells (supplemental Methods). Hierarchical clustering revealed similarity in the expression pattern of TgERG leukemias and lin– cells compared with samples taken from the whole bone marrow of WT animals (Figure 1F), confirming the early progenitor phenotype of TgERG leukemias. In addition, gene set enrichment analysis (GSEA) revealed significant enrichment of the TgERG leukemia gene signature in the expression profile of MEPs compared with other myeloid progenitors19 (Figure 1G; supplemental Figure 2), providing further support for the MEP phenotype of these leukemias.
We next wanted to determine the relevance of TgERG myeloid leukemias to the human disease. Using GSEA, we found significant enrichment of high ERG–expressing human AML signature9 in the gene expression profile of TgERG leukemia compared with the WT control (Figure 1H). Because ERG was recently found to be part of a gene signature of human AML stem cells,11 we performed GSEA using expression data of AML cases20 and found enrichment of a mouse TgERG expression signature in human AML stem and progenitor cells (Figure 1I; supplemental Figure 3). Recently, Diffner et al showed that the ERG promoter and stem-cell enhancer activate gene expression signatures that are associated with clinical outcome of human AML.21 This signature is also significantly enriched in TgERG leukemia, demonstrating the similarity in transcriptional programs of mouse and human AML (Figure 1J). Together these results establish that TgERG leukemia carries an early stem/progenitor gene signature and is similar to human leukemias with high expression of ERG and/or a stem cell signature.
ChIP-Seq of ERG in Tg leukemias directly implicates ERG in leukemogenic transcriptional programs
We next wanted to investigate how ectopic ERG expression activates leukemogenic transcriptional programs. We used ChIP-Seq to generate genome-scale catalogs of sequences bound by ERG in primary cells from 2 leukemic mice (Tg1 and Tg2). Before sequencing, ChIP material was validated by quantitative RT-PCR. We have previously identified an enhancer in the first intron of Hhex/Prh (+1-kb enhancer)25 that is strongly bound by LMO2, FLI1, and ERG in human T-ALL cells.26 Significant binding of ERG to this site compared with an IgG control sample was observed in both primary mouse TgERG leukemias (Figure 2A). The ChIP material was then analyzed by high-throughput sequencing, yielding ∼28 million uniquely mappable reads for both samples. Visualization of the raw data across the Hhex locus reproduced the PCR results with specific enrichment of the Hhex +1 region (Figure 2B). Binding peaks were also seen in gene loci of other transcription factors important for hematopoietic progenitor and/or megakaryocyte development such as Nfe2 and Runx1 (Figure 2B).
ERG ChIP-Seq in TgERG mice. (A) Validation of ChIP quality by quantitative RT-PCR of the Hhex +1-Kb enhancer-fold enrichment compared with the IgG control in 2 TgERG leukemias. (B) Density plots visualizing ERG binding in 3 known hematopoietic target gene loci, Hhex, Nfe2, and Runx-1. (C) Venn diagram intersecting ERG peaks in the 2 Tg leukemias shows marked similarity between 2 samples. (D) Pie chart showing ERG distribution across genomic regions using the designated high-confidence peaks. (E) De novo motif discovery performed on ERG high-confidence peaks identified ETS and GATA motifs as highly enriched. (F) Biological function of ERG-bound genes as identified by the Genomic Regions Enrichment of Annotations Tool. A table summarizing the most enriched mouse phenotypes is presented and phenotypes related to mega-erythro lineage are highlighted in yellow.
ERG ChIP-Seq in TgERG mice. (A) Validation of ChIP quality by quantitative RT-PCR of the Hhex +1-Kb enhancer-fold enrichment compared with the IgG control in 2 TgERG leukemias. (B) Density plots visualizing ERG binding in 3 known hematopoietic target gene loci, Hhex, Nfe2, and Runx-1. (C) Venn diagram intersecting ERG peaks in the 2 Tg leukemias shows marked similarity between 2 samples. (D) Pie chart showing ERG distribution across genomic regions using the designated high-confidence peaks. (E) De novo motif discovery performed on ERG high-confidence peaks identified ETS and GATA motifs as highly enriched. (F) Biological function of ERG-bound genes as identified by the Genomic Regions Enrichment of Annotations Tool. A table summarizing the most enriched mouse phenotypes is presented and phenotypes related to mega-erythro lineage are highlighted in yellow.
Elimination of artifactual peaks found in both test and control IgG ChIP samples yielded 5738 ERG-specific peaks in the Tg1 sample and 3751 peaks in the Tg2 sample. Eighty-five percent of the peaks detected in the Tg2 leukemia sample were also detected in Tg1 leukemia (Figure 2C), confirming that ERG binding was very similar in both. We therefore performed all subsequent analysis on the 3201 regions bound by ERG in both samples. Of these high-confidence regions, 59% locate to promoters, 23% to intragenic, and 18% to intergenic regions, respectively (Figure 2D).
De novo motif discovery confirmed the high quality of this dataset by recovering an ETS consensus binding motif (Figure 2Ei) as the most overrepresented motif. Of note, the GATA consensus motif (Figure 2Eii) was also highly enriched, which may be related to the known cooperative DNA binding of the ERG paralogue, FLI1, with GATA1 in megakaryocyte-specific promoters and with Gata2 in blood stem-cell enhancers.27 We next used the “Genomic Regions Enrichment of Annotations Tool” (GREAT28 ) to identify over-represented biological functions for genes next to the 3201 regions bound by ERG in both leukemia samples. Of note, all of the top 10 ranked mouse phenotypes were hematopoietic, including 8 megakaryocyte or red cell–related phenotypes (Figure 2F). This analysis therefore demonstrated that ERG regulates a distinct transcriptional program in the leukemic cells of the mega-erythroid lineage.
Integrated genomic analysis reveals an ERG-regulated transcriptional program characteristic of human leukemia progenitors
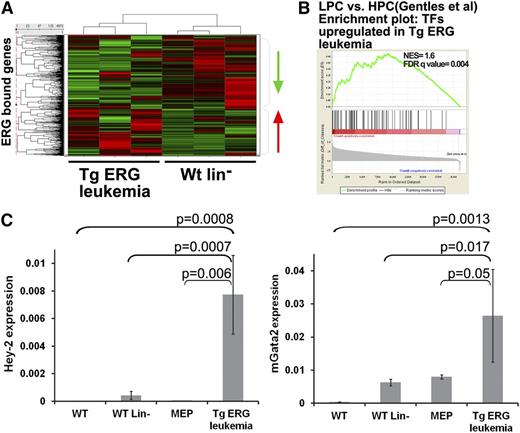
To identify candidate ERG targets, the high confidence ERG peaks were mapped to 3433 genes. We next integrated the ChIP-Seq and expression data by matching all ERG-bound genes to the expression data and creating expression profiles of TgERG leukemia compared with WT lin– cells (Figure 3A). Unsupervised clustering based on expression data for the ERG candidate target genes partitioned the samples into leukemic and nonleukemic cell types, consistent with the notion that elevated ERG drives expression changes relevant to the disease phenotype. The DAVID functional annotations tool29 suggests that ERG directly regulates the expression of a large group of transcription regulators in TgERG leukemia compared with WT lin– cells (supplemental Table 1), and GSEA confirmed that the upregulated transcription factors are enriched in human AML progenitors compared with normal hematopoietic progenitors (Figure 3B). Thus, ERG directly activates a series of transcription factors that characterize human AML progenitors.
Integration of ChIP-Seq and expression data reveals transcriptional programs regulated by ERG in Tg leukemia. (A) Heat map depicting gene expression of ERG-bound genes in TgERG leukemia compared with the WT lin– controls. (B) GSEA using upregulated transcription factors (TFs) in Tg-ERG leukemia shows significant enrichment in human AML progenitors (LPC) compared with normal hematopoietic progenitors (HPC20 ). (C) Validation of Gata2 and Hey2 upregulation in TgERG leukemia compared with WT bone marrow, WT lin– bone marrow, and WT MEPs. Quantitative RT-PCR results normalized to β-actin are shown (mean ± SEM, n = 3 for WT, WT lin–, and WT MEPs; n = 5 for TgERG leukemia).
Integration of ChIP-Seq and expression data reveals transcriptional programs regulated by ERG in Tg leukemia. (A) Heat map depicting gene expression of ERG-bound genes in TgERG leukemia compared with the WT lin– controls. (B) GSEA using upregulated transcription factors (TFs) in Tg-ERG leukemia shows significant enrichment in human AML progenitors (LPC) compared with normal hematopoietic progenitors (HPC20 ). (C) Validation of Gata2 and Hey2 upregulation in TgERG leukemia compared with WT bone marrow, WT lin– bone marrow, and WT MEPs. Quantitative RT-PCR results normalized to β-actin are shown (mean ± SEM, n = 3 for WT, WT lin–, and WT MEPs; n = 5 for TgERG leukemia).
Quantitative RT-PCR validation confirmed that Gata2 and Hey2 (hairy/enhancer-of-split related with YRPW motif 2) are highly upregulated transcription factors in TgERG leukemia (Figure 3C). Importantly, this upregulation was significant also when compared with WT MEPs, suggesting a role for these transcription factors specifically in ERG-driven leukemia and not simply as markers of this progenitor cell (Figure 3C). GATA2 is an important regulator early in hematopoiesis and is perturbed in hematologic malignancies.30-32 Moreover, ERG was shown to be a direct upstream regulator of GATA2 during the second wave of hematopoiesis.2 Involvement of the βHLH transcription factor Hey2 in hematopoietic stem cell specification in zebrafish has been recently suggested.33 These results coupled with the upregulation of both Hey2 and Gata2 in AML progenitor cells20 indicate that ERG overexpression induces a transcriptional program that maintains the immature nature of ERG-driven leukemia.
Activation of RAS in TgERG leukemia
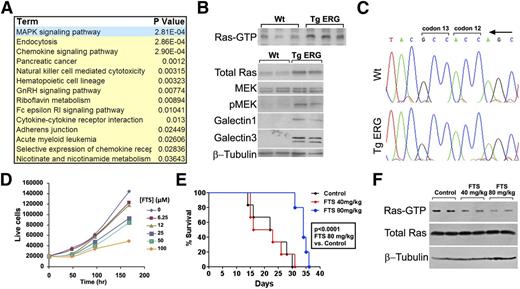
Analysis of direct ERG targets through an integrated expression array and ChIP-Seq profiling allowed us to identify signaling pathways that are likely key players in ERG-driven myeloid leukemia. Using the DAVID functional annotations tool, we found upregulation of several cancer-related signaling pathways in the Tg leukemia, with the MAPK pathway being the most enriched (Figure 4A). Quantitative RT-PCR confirmed elevated expression of MAPK pathway genes such as the dual-specificity kinase, Map2k3, the transforming growth factors TGFβ1 and TGFα, and the growth factor receptor binding protein Grb2 (supplemental Figure 4), which is known to recruit guanine nucleotide exchange factors that facilitate activation of the small GTPase RAS.34 Accordingly, RAS is activated in TgERG leukemias as depicted by elevated levels of active Ras-GTP and its downstream effectors (Figure 4B). In AML, RAS is frequently activated by somatic mutation occurring in codons 12, 13, and 63.35 However, no such mutations were found in TgERG leukemia in the 3 RAS isoforms: H, K, or N-RAS (Figure 4C), confirming the activation of the pathway by an upstream transcriptional program.
The Ras pathway is activated in TgERG leukemia. (A) Table summarizing most upregulated pathways in TgERG leukemia compared with WT lin– cells according to the DAVID functional annotations tool. (B) Analysis of the Ras pathway in protein extracts from the spleens of TgERG mice or WT littermates: Ras-GTP pull-down to visualize the active Ras state (upper panel) and Ras pathway protein expression by Western blotting (lower panel). (C) Absence of mutations in Ras in cDNA prepared from TgERG leukemia cells: representative chromatograms showing K-Ras codons 12 and 13. (D) Growth curve of TgERG leukemia cells treated with escalating doses of FTS (mean ± SEM, n = 3; experiment repeated 3 times). (E) Kaplan-Meier survival curve of NSG mice transplanted with TgERG leukemia cells and treated with FTS by daily gavage starting 4 days after transplantation. FTS treatment at 80 mg/kg significantly prolonged survival of leukemia-bearing NSG mice (median survival = 22.5 and 34 days for mice receiving vehicle or FTS 80 mg/kg, respectively, log-rank P < .0001, n = 6). (F) Depletion of Ras-GTP in mice treated with FTS 80 mg/kg. Protein extracts were prepared from the spleens of 2 NSG mice receiving vehicle control, 2 mice receiving FTS 40 mg/kg, and 2 mice receiving 80 mg/kg. Extracts were used for either Ras-GTP pull-down to detect active Ras levels or for Western blotting to detect total Ras and β-tubulin levels.
The Ras pathway is activated in TgERG leukemia. (A) Table summarizing most upregulated pathways in TgERG leukemia compared with WT lin– cells according to the DAVID functional annotations tool. (B) Analysis of the Ras pathway in protein extracts from the spleens of TgERG mice or WT littermates: Ras-GTP pull-down to visualize the active Ras state (upper panel) and Ras pathway protein expression by Western blotting (lower panel). (C) Absence of mutations in Ras in cDNA prepared from TgERG leukemia cells: representative chromatograms showing K-Ras codons 12 and 13. (D) Growth curve of TgERG leukemia cells treated with escalating doses of FTS (mean ± SEM, n = 3; experiment repeated 3 times). (E) Kaplan-Meier survival curve of NSG mice transplanted with TgERG leukemia cells and treated with FTS by daily gavage starting 4 days after transplantation. FTS treatment at 80 mg/kg significantly prolonged survival of leukemia-bearing NSG mice (median survival = 22.5 and 34 days for mice receiving vehicle or FTS 80 mg/kg, respectively, log-rank P < .0001, n = 6). (F) Depletion of Ras-GTP in mice treated with FTS 80 mg/kg. Protein extracts were prepared from the spleens of 2 NSG mice receiving vehicle control, 2 mice receiving FTS 40 mg/kg, and 2 mice receiving 80 mg/kg. Extracts were used for either Ras-GTP pull-down to detect active Ras levels or for Western blotting to detect total Ras and β-tubulin levels.
To evaluate the role of Ras in TgERG leukemia cells, we used a specific Ras inhibitor, S-trans, trans FTS (Salirasib). FTS competes with RAS on its membrane anchorage sites, leading to dislodgement and degradation of the protein.18 FTS-mediated Ras inhibition disrupts tumor growth in various cancer models,18,36,37 and clinical trials for several cancers38 (ClinicalTrials.gov; NCT00867230, NCT00531401) have demonstrated its clinical activity and safety. FTS caused a dose- and time-dependent growth inhibition of TgERG leukemia cells (Figure 4D) and prolonged the survival of NSG mice transplanted with TgERG leukemia cells (Figure 4E) without affecting mouse red blood cells and platelet counts (supplemental Figure 5). The effective FTS dose also caused a significant decrease in active Ras-GTP levels in leukemia-infiltrated spleens (Figure 4F). These results indicate that RAS inhibition may serve as a therapeutic approach in AML patients expressing high levels of ERG.
Pim1 kinase is a direct ERG target activated through a previously unknown +10 enhancer
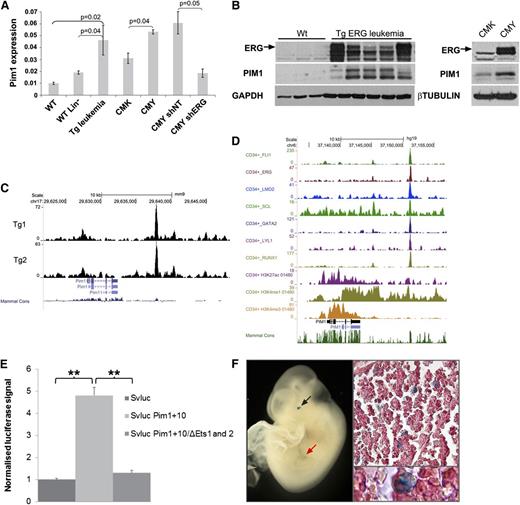
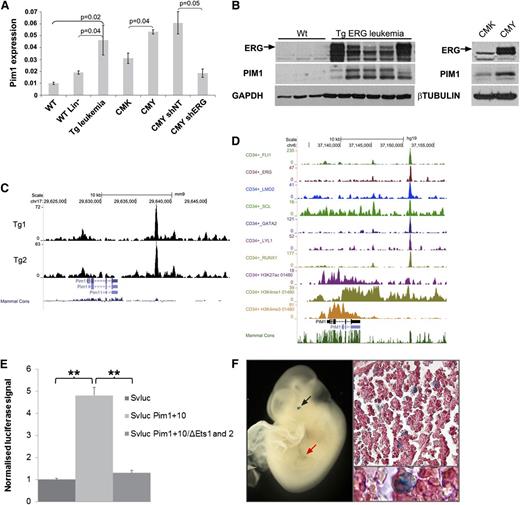
As shown in Figure 4A, integrative genomic analysis of direct ERG targets revealed upregulation of AML-related genes. One of these genes, Pim1, encodes an oncogenic serine/threonine kinase.39 We confirmed increased expression of Pim1 RNA and protein in independent TgERG mouse samples as well as in the human AMKL cell line CMY, in which ERG is highly expressed (Figure 5A-B). PIM1 levels were lower in CMK cells, which express lower levels of ERG and upon knockdown of ERG in CMY cells (Figure 5A-B), confirming that ERG regulates PIM1 expression in leukemia.
Pim1 is a direct ERG target activated by a +10 enhancer site. (A) Pim1 levels by quantitative RT-PCR in TgERG leukemia and mouse WT whole or lineage-negative bone marrow, in human CMK (low ERG) or CMY (high ERG) cells, and in CMY cells transfected with either nontargeting shRNA (shNT) or ERG (shERG) shRNA. (B, left panel) ERG and Pim1 protein levels in the bone marrow of TgERG leukemic mice and WT littermates; (right panel) ERG and PIM1 protein levels in CMK and CMY human AML cells. (C) Density plots showing ERG binding to Pim1 loci in 2 TgERG leukemias. (D) Density plots showing occupancy of the PIM1 loci in human CD34+ cells by a heptad of transcription factors and by open chromatin marks. (E) Luciferase reporter assay. The Pim1 +10 candidate regulatory region was cloned into a luciferase reporter plasmid and stably transfected to L8057 cells. Cells expressing the Pim1 +10 region showed a four to sixfold increase in promoter activity compared with the empty vector. Mutations in the 2 ETS motifs in the Pim1 +10 region nearly abolished its enhancing effect. (F) Transgenic reporter mice: The Pim1 +10 region was cloned into a lacZ reporter vector and used to generate transgenic F0 embryos. Typical images showing Xgal staining in fetal liver of E10.5 embryo in either whole embryo (left) or in fetal liver sections (right).
Pim1 is a direct ERG target activated by a +10 enhancer site. (A) Pim1 levels by quantitative RT-PCR in TgERG leukemia and mouse WT whole or lineage-negative bone marrow, in human CMK (low ERG) or CMY (high ERG) cells, and in CMY cells transfected with either nontargeting shRNA (shNT) or ERG (shERG) shRNA. (B, left panel) ERG and Pim1 protein levels in the bone marrow of TgERG leukemic mice and WT littermates; (right panel) ERG and PIM1 protein levels in CMK and CMY human AML cells. (C) Density plots showing ERG binding to Pim1 loci in 2 TgERG leukemias. (D) Density plots showing occupancy of the PIM1 loci in human CD34+ cells by a heptad of transcription factors and by open chromatin marks. (E) Luciferase reporter assay. The Pim1 +10 candidate regulatory region was cloned into a luciferase reporter plasmid and stably transfected to L8057 cells. Cells expressing the Pim1 +10 region showed a four to sixfold increase in promoter activity compared with the empty vector. Mutations in the 2 ETS motifs in the Pim1 +10 region nearly abolished its enhancing effect. (F) Transgenic reporter mice: The Pim1 +10 region was cloned into a lacZ reporter vector and used to generate transgenic F0 embryos. Typical images showing Xgal staining in fetal liver of E10.5 embryo in either whole embryo (left) or in fetal liver sections (right).
Magistroni et al recently showed that ERG upregulates PIM1 in prostate cancer cells via direct binding to the PIM1 promoter.40 Analysis of our ERG ChIP-Seq data in TgERG leukemias revealed weak binding to the Pim1 promoter but a much stronger ERG peak about 10 kb downstream of the promoter (Figure 5C). This region contains several evolutionarily conserved transcription factor consensus binding motifs including ETS motifs (supplemental Figure 6), suggesting the presence of an enhancer element. Notably, the corresponding PIM1 +13 region in human CD34+ cells is bound by a heptad of transcription factors, including ERG, known to activate stem cell programs in AML,21 and it displays open chromatin marks (Figure 5D). In addition, ERG binds the PIM1 +13 region in the human AML cell lines, KG-1 and ME-1 (supplemental Figure 7).
To test whether this region has enhancer activity, a DNA fragment corresponding to the ERG-bound region (+10-kb region) was inserted downstream of a minimal promoter/luciferase reporter cassette and assayed after stable transfection in the murine megakaryoblastic cell line L8057.15 Inclusion of the Pim1 +10-kb region caused a four- to sixfold increase of promoter activity, which was almost completely abolished when the 2 conserved ETS motifs were mutated (Figure 5E). To confirm bonafide enhancer activity of the Pim1 +10 region, we generated transgenic mouse embryos with a corresponding lacZ reporter construct. Analysis of E11.5 midgestation embryos confirmed in vivo activity in fetal liver hematopoietic cells, some of which displayed megakaryocyte-like characteristics (Figure 5F). The enhancer element also directed expression to the eye, which reflects Pim1 known expression in the mouse embryonic neural retina.41 Taken together, these data suggest that ERG expression in the Tg leukemias mediates elevated levels of Pim1 through a previously unknown Pim1 3′ enhancer.
Pim1 kinase is a potential therapeutic target in ERG-expressing AML
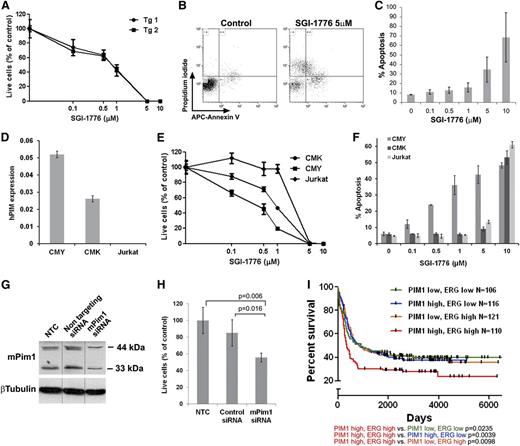
PIM inhibitors have been evaluated in both preclinical and clinical trials for hematologic malignancies42,43 (ClinicalTrials.gov; NCT00848601, NCT01239108). To test Pim1 as a potential therapeutic target of high ERG-expressing leukemias we treated mouse TgERG leukemia cells and human AMKL cells with the PIM1 inhibitor SGI-1776.42,43 SGI-1776 markedly inhibited TgERG leukemia cell growth and induced apoptosis in a dose-dependent manner (Figure 6A-C). Similar results were obtained with the human AMKL cell lines, CMK and CMY (Figure 6E-F). Moreover, CMY cells were significantly more sensitive, exhibiting an IC50 of 512nM compared with 923nM in CMK cells, and also exhibiting increased apoptosis (Figure 6E-F). This difference correlated well with the higher levels of PIM1 in CMY cells (which also express higher levels of ERG) (Figure 6D). In contrast, Jurkat cells that do not express PIM1 (Figure 6D) were resistant to SGI-1776 (Figure 6E-F). Importantly, the specificity of the observed SGI-1776 growth inhibitory effects is further supported by the growth inhibition of the TgERG leukemia cell line after siRNA-mediated knockdown of PIM1 (Figure 6G-H). The significance of the ERG-PIM1 axis is further supported by clinical data; in AML patients classified according to the levels of ERG and PIM1 expression (supplemental Figure 8), high expression of both genes is significantly associated with decreased survival rates (Figure 6I). Thus, PIM1 may be a highly relevant therapeutic target in these leukemias.
The PIM1 inhibitor, SGI-1776, inhibits proliferation and induces apoptosis of leukemia cells. (A) Survival curve of TgERG leukemia cells of 2 TgERG mice treated with escalating doses of SGI-1776 for 3 days (mean ± SEM, n = 4). (B-C) Dose-dependent apoptosis of TgERG leukemia cells treated with SGI-1776 for 24 hours. (B) Flow cytometry analysis of cells stained with Annexin V and propidium iodide 24 hours after treatment with either vehicle or 5 µM SGI-1776. (C) Summary of the percentage of Annexin V–positive cells as a function of SGI-1776 dose (mean ± SEM, n = 3) (D) PIM1 expression levels by quantitative RT-PCR (normalized to β-actin) in human leukemia cell lines expressing high (CMY) low (CMK) or no ERG (Jurkat, mean ± SEM, n = 3). (E) Survival curve of CMK, CMY, and Jurkat cells treated with escalating doses of SGI-1776 for 2 days (mean ± SEM, n = 4, 1-way analysis of variance P = .026). (F) Percentage of Annexin V–positive CMK, CMY, and Jurkat cells treated for 24 hours with escalating doses of SGI-1776 (mean ± SEM, n = 3). (G-H) Pim1 knockdown inhibits growth of TgERG leukemia cells. TgERG leukemia cells were transfected with Pim1 siRNA and 3 days later collected and used for either protein extraction to determine PIM1 levels (G, vertical lines indicate repositioned gel lanes) or counted to determine live cell number (H, mean ± SEM, n = 3). (I) Combined high expression of ERG and PIM1 confers a poor prognosis in AML patients. Kaplan-Meier plots comparing survival in 4 groups of human AML44 classified by PIM1 and ERG expression (see supplemental Figure 8). Significance levels (log-rank P values) are given with respect to the PIM1/ERG high subpopulation.
The PIM1 inhibitor, SGI-1776, inhibits proliferation and induces apoptosis of leukemia cells. (A) Survival curve of TgERG leukemia cells of 2 TgERG mice treated with escalating doses of SGI-1776 for 3 days (mean ± SEM, n = 4). (B-C) Dose-dependent apoptosis of TgERG leukemia cells treated with SGI-1776 for 24 hours. (B) Flow cytometry analysis of cells stained with Annexin V and propidium iodide 24 hours after treatment with either vehicle or 5 µM SGI-1776. (C) Summary of the percentage of Annexin V–positive cells as a function of SGI-1776 dose (mean ± SEM, n = 3) (D) PIM1 expression levels by quantitative RT-PCR (normalized to β-actin) in human leukemia cell lines expressing high (CMY) low (CMK) or no ERG (Jurkat, mean ± SEM, n = 3). (E) Survival curve of CMK, CMY, and Jurkat cells treated with escalating doses of SGI-1776 for 2 days (mean ± SEM, n = 4, 1-way analysis of variance P = .026). (F) Percentage of Annexin V–positive CMK, CMY, and Jurkat cells treated for 24 hours with escalating doses of SGI-1776 (mean ± SEM, n = 3). (G-H) Pim1 knockdown inhibits growth of TgERG leukemia cells. TgERG leukemia cells were transfected with Pim1 siRNA and 3 days later collected and used for either protein extraction to determine PIM1 levels (G, vertical lines indicate repositioned gel lanes) or counted to determine live cell number (H, mean ± SEM, n = 3). (I) Combined high expression of ERG and PIM1 confers a poor prognosis in AML patients. Kaplan-Meier plots comparing survival in 4 groups of human AML44 classified by PIM1 and ERG expression (see supplemental Figure 8). Significance levels (log-rank P values) are given with respect to the PIM1/ERG high subpopulation.
Discussion
The ETS transcription factor ERG is an independent prognostic factor in AML and T-ALL,8-10 yet it is unclear whether its expression is a marker of the early differentiation block of the leukemic cells or whether ERG actively contributes to the leukemic phenotype. Although we and others7,45 have previously shown that ERG is a megakaryocytic oncogene in vivo, these transduction-transplantation experiments were associated with extremely high levels of ERG expression and an immunophenotype that is not found in human AML with high ERG expression. We show here that transgenic expression of the human ERG gene causes leukemias in mice that are highly similar to human leukemias. Integration of ChIP-Seq and gene expression data demonstrates that ERG activates a transcriptional program characteristic of leukemia stem and progenitor cells. ERG also induces the expression of the oncogenic kinase PIM1 through a novel 3′ enhancer. This and the indirect activation of the Ras pathway therefore emerge as potential therapeutic targets of ERG-driven leukemias.
ERG is a member of a prognostically significant human leukemia stem cell gene signature.11 Enrichment of a stem/progenitor signature in our Tg-ERG mouse AML suggests that ERG may not simply be a marker but instead constitutes an important driver of the leukemia stem cell signature. Of note, direct ERG target genes in the TgERG leukemia include transcription factors such as GATA2 and HEY2, with GATA motifs also being highly enriched within the regions bound by ERG. GATA2 is a key regulator of hematopoietic stem cell32 and megakaryocyte development,46 and high GATA2 expression has recently been reported as a poor prognostic factor in pediatric AML.31 HEY2 is largely recognized as a regulator of cardiovascular development47 but was shown to act upstream of Notch to induce hematopoietic stem cell formation in zebrafish embryos.33 The involvement of both HEY2 and GATA2 in early hematopoiesis, their increased expression in human AML progenitors,20 and their overexpression specifically in TgERG myeloid but not lymphoid leukemia (supplemental Figure 9) suggest a role for both transcription factors in sustaining the immature nature of ERG-induced myeloid leukemia.
When comparing ERG-bound genes in the Tg leukemia and in mouse hematopoietic progenitor cells,3 we found a significant overlap between the 2 groups (supplemental Figure 10), suggesting similar transcriptional programs. However, the enrichment of ERG on promoters is intriguing because in hematopoietic cells, the minority of peaks are in promoter regions.3 One could speculate that this binding amplifies the cellular state similar to promoter binding by c-Myc in cancer cells. Rather than binding a new set of genes, c-Myc has been suggested to amplify the output of existing gene expression programs and thereby reduce rate-limiting constraints for tumor cell growth and proliferation.48
Interestingly, we found that the most downregulated pathways in TgERG leukemia are purine and pyrimidine metabolism (supplemental Table 2). Nucleotide deficiency has recently been shown to promote genomic instability, a key factor in the first stages of cancer formation.49 Therefore, it is possible that overexpression of ERG in hematopoietic cells contributes to genomic instability, which in conjunction with other affected pathways, may drive the leukemogenic process.
A major goal of this work was to identify ERG targets that may be pharmaceutically targeted. The most enriched signaling pathway in TgERG leukemia is the MAPK pathway, a pathway known to be downstream of the small GTPase, RAS, which is indeed activated in Tg leukemia (Figure 4B). Ras is a well-known oncogene and is constitutively activated by common somatic mutations in ∼20% of AML cases.35 Consistent with our findings of transcriptional activation, we found no activating mutations in Ras, but we did observe upregulation of Grb2, a known activator of Ras.34 The observed growth inhibitory activity of FTS in ERG-expressing leukemia cells in vitro and in vivo establish the Ras pathway as a promising target for AML therapeutics, not only for those 20% of AML cases with RAS mutations but also for high ERG–expressing AML.
We also identified PIM1 kinase as a direct ERG target activated through a novel enhancer, which we further characterized in mouse and human cells. The PIM1 serine/threonine kinase, first described as a proviral integration site in murine T-cell lymphomas,50 is frequently overexpressed in various malignancies including hematologic51 and prostate cancers.52 In AML, PIM1 regulates cell proliferation and survival as well as homing and migration.43,53,54 We show the prognostic relevance of PIM1 upregulation in high ERG–expressing AML by the significantly decreased survival of these patients. We therefore explored the effects of Pim1 inhibition in TgERG leukemia by a PIM inhibitor, SGI-1776.43 SGI-1776 has been shown to inhibit growth and promote apoptosis of hematopoietic and prostate cancer cells.43,55 It is also a potent inhibitor of FLT3 and TRKA, neither of which is expressed in TgERG leukemia (not shown). SGI-1776 treatment inhibited growth and induced apoptosis of TgERG leukemia cells as well as in human leukemia cells, where the inhibitor efficacy was proportional to PIM1 expression levels. siRNA-mediated Pim1 knockdown also inhibited the growth of TgERG leukemia cells, verifying the specificity of SGI-1776 to Pim1. These results indicate that PIM1 inhibition should be considered for therapy of high ERG–expressing AML. SGI-1776 has already been evaluated in phase 1 clinical trials for Refractory Prostate Cancer, Relapsed/Refractory Non-Hodgkin’s Lymphoma and Relapsed/Refractory Leukemias (ClinicalTrials.gov; NCT00848601, NCT01239108). Although these trials were terminated because of dose-limiting toxicity, the beneficial effect of PIM1 inhibition in AML preclinical models has been established.42,43
The striking observation that high PIM1 expression is associated with poor prognosis in the context of high ERG expression has implication for the future design of clinical trials. Targeting PIM1 activity may be relevant in only a subset of AMLs with high ERG expression or stem cell characteristics. We believe that measuring ERG and PIM1 expression as biomarkers should be embedded in future clinical trials associated with PIM1 inhibitors. Interestingly, combined treatment of TgERG cells with both PIM and Ras inhibitors yielded an additive inhibitory effect on cell proliferation (supplemental Figure 11), indicating that targeting these 2 pathways may improve therapeutic effects in ERG-driven AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We appreciate the assistance of Drs Yoram Groner and Ditsa Levanon in generation of the transgenic mice. We are indebted to Dr Mali Salmon for advice, and assistance by members of the Izraeli, Gottgens, Pimanda, and Aplan laboratories.
This work was supported in part by the Israel Science Foundation, Israel Centers of Research Excellence 41/11, Children with Cancer UK and the Waxman Foundation (S.I.), the Israel Cancer Research Foundation (L.G.), a Daniel Turnberg UK/Middle East Travel Fellowship from the Academy of Medical Sciences (L.G.), The European Hematology Association (Y.B.), Marie Curie Intra-European Fellowship 237296 (M.R.T.), a Leukaemia and Lymphoma Research Studentship (J.S.), programmatic funding from Leukaemia and Lymphoma Research and Cancer Research UK (B.G.), and core support grants by the Wellcome Trust to the Cambridge Institute for Medical Research and Wellcome Trust–MRC Cambridge Stem Cell Institute (B.G.). This work was also supported in part by Grant NCI P50 CA 140158 and The Coleman Leukemia Research Foundation (C.D.B.).
Authorship
Contribution: L.G., J.E.P., B.G., and S.I. designed the study; L.G., M.R.T., D.B., J.J.-H., R.L.H., and D.B. analyzed the data; L.G., M.R.T., Y.B., S.J.K., J.S., K.K., G.S., J.J.-H., and A.B. performed experiments; Y.K., G.M., C.D.B., and P.D.A. provided research guidance; and L.G., M.R.T., J.E.P., B.G., and S.I. helped write the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shai Izraeli, Cancer Research Center, Sheba Medical Center, Tel Hashomer, Ramat Gan, 52621, Israel and Department of Human Molecular Genetics and Biochemistry, Tel Aviv University, Tel Aviv, 69978, Israel; e-mail: Shai.Izraeli@sheba.health.gov.il; and Berthold Göttgens, Cambridge Institute for Medical Research and Wellcome Trust and MRC Cambridge Stem Cell Institute, Hills Road, Cambridge CB2 0XY, UK; e-mail: bg200@cam.ac.uk.
References
Author notes
L.G. and M.R.T. contributed equally to this study.