Key Points
SF-mutant myeloid malignancies transcend the boundaries between AML and MDS.
Integrated analysis of gene expression and DNA-methylation profiles in leukemia uncovers novel subtypes.
Abstract
Mutations in splice factor (SF) genes occur more frequently in myelodysplastic syndromes (MDS) than in acute myeloid leukemias (AML). We sequenced complementary DNA from bone marrow of 47 refractory anemia with excess blasts (RAEB) patients, 29 AML cases with low marrow blast cell count, and 325 other AML patients and determined the presence of SF-hotspot mutations in SF3B1, U2AF35, and SRSF2. SF mutations were found in 10 RAEB, 12 AML cases with low marrow blast cell count, and 25 other AML cases. Our study provides evidence that SF-mutant RAEB and SF-mutant AML are clinically, cytologically, and molecularly highly similar. An integrated analysis of genomewide messenger RNA (mRNA) expression profiling and DNA-methylation profiling data revealed 2 unique patient clusters highly enriched for SF-mutant RAEB/AML. The combined genomewide mRNA expression profiling/DNA-methylation profiling signatures revealed 1 SF-mutant patient cluster with an erythroid signature. The other SF-mutant patient cluster was enriched for NRAS/KRAS mutations and showed an inferior survival. We conclude that SF-mutant RAEB/AML constitutes a related disorder overriding the artificial separation between AML and MDS, and that SF-mutant RAEB/AML is composed of 2 molecularly and clinically distinct subgroups. We conclude that SF-mutant disorders should be considered as myeloid malignancies that transcend the boundaries of AML and MDS.
Introduction
Myelodysplastic syndromes (MDS) are characterized by a deregulation of blood cell formation and frequently develop into acute myeloid leukemia (AML). MDS patients feature recurrent somatic mutations in multiple components of the RNA splicing machinery.1-4 These mutations are frequently seen in the splice factor (SF) genes SRSF2, U2AF35, ZRSR2, U2AF65, SF1, SF3B1, SF3A1, or PRPF40B.1-3 Among the many nonrecurrent missense mutations, 8 mutational hotspots were found, ie, in U2AF35 (2 hotspots), SRSF2 (1 hotspot), and SF3B1 (5 hotspots).1 Although these SF mutations have been reported to frequently associate with the presence of ring sideroblasts (RS),1,5 MDS without RS can harbor SF mutations as well.1,2 Mutations in SF3B1 are associated with refractory anemia with ring sideroblasts, whereas in MDS without RS no association with a specific mutation was observed.1 Some of the SF mutations also appeared to have prognostic relevance.6-9
In the past, the French-American-British classification for MDS and AML was applied to delineate the transitional zone in marrow and blood blast percentages that separate MDS and AML. Refractory anemia with excess blasts (RAEB) was defined as an MDS with ≥5% but ≤20% blasts in the bone marrow (BM), whereas RAEB in transformation was designated to cases with BM blasts >20% but <30%, with ≥5% blasts in the blood or the presence of Auer rods.10,11 Blast percentages are still applied to discriminate RAEB patients that are considered to be MDS subtypes closely related to AML, whereas RAEB in transformation is now classified in most cases as AML according to the World Health Organization.12 This means that the decision to classify RAEB cases as MDS is relatively arbitrary, and molecular abnormalities have not been included for the discrimination of AML and MDS. It is therefore possible that subsets of RAEB and AML patients, classified according to the World Health Organization, belong to the same molecular class of abnormalities. We hypothesized that SF mutations in RAEB and AML may reveal unique molecular leukemia subtypes at the boundary of MDS and AML. To address this issue, in this study we investigated the distribution of 8 hotspot SF gene mutations1 in RAEB (n = 47) and in 354 AML cases. Within the AML group, we separated AMLs with a low blast cell count (AML-LBC) (>20% but <30%, n = 29) from the other AML cases (n = 325). We show that RAEB, AML-LBC, and AML cases with SF mutations share highly similar phenotypes and suggest that these malignancies should be considered as 1 typical SF-mutant leukemia subset.
AML subtypes with unique molecular defects, such as patients with recurrent chromosomal translocations t(8;21), t(15;17), or inv(16) or with mutations in CEBPA or in NPM1, can be uncovered very specifically using gene expression profiling or DNA-methylation profiling (genomewide messenger RNA [mRNA] expression profiling ([GEP]13 or DNA-methylation profiling [DMP]14 ) data, derived from large AML patient cohorts.13,15-17 Application of GEP or DMP in cohorts that also included RAEB, AML-LBC, and AMLs did not reveal distinctive gene expression or methylation patterns for these malignancies.13 14 We hypothesize that SF-mutant myeloid disorders constitute a biological entity with distinct gene expression and methylation patterns. To address this hypothesis, we developed an approach toward integrative analysis of the GEP and DMP data sets. Our data point to the existence of 2 SF-mutant clusters, each with a different GEP/DMP signature with distinct additional molecular features and response to treatment.
Material and methods
Patients and molecular analyses
Diagnostic BM or peripheral blood samples from 344 adults were analyzed; patients were enrolled on Haemato Oncology Foundation for Adults in the Netherlands/Swiss Group for Clinical and Epidemiological Cancer Research protocols 04, 04A, 29, 32, 42, and 43 (available at www.hovon.nl).18-20 Patients provided written informed consent in accordance with the Declaration of Helsinki, and all trials were approved by the Institutional Review Board of Erasmus University Medical Center. Among the AML patients that were found in the 2 newly identified clusters (ie, cluster 3 or cluster 11), 1 case (6453) had a known antecedent MDS before inclusion in this study. Mutational analyses in NPM1, FLT3, CEBPA IDH1, DNMT3A, NRAS, KRAS, and ASXL1 were carried out as described previously.15,21-23 Summary of clinical, (cyto)genetical, and molecular features of the patients have been described previously.14 Mutation analyses for the genes U2AF35, SRSF2, and SF3B1 were performed by denaturing high-performance liquid chromatography (dHPLC). Sanger sequencing was subsequently performed on samples with an abnormal dHPLC profile using the primer sets as shown in supplemental Table 1 on the Blood Web site. RNA and complementary DNA synthesis was performed as previously described.13 Whole-exome sequencing (WES) has been performed on DNA isolated from RAEB, AML-LBC, or AML blasts (14 patient samples in total) purified by Ficoll-Hypaque (Nygaard) centrifugation, and cryopreserved in aliquots.24 CD3+ T cells were expanded from diagnostic BM or peripheral blood specimens and used as controls for WES to determine acquired mutations in AML blasts. Primary cells were seeded in supplemented RPMI (10% fetal calf serum/100 U/mL penicillin/streptomycin) at ∼1 × 106/mL in a 48-well plate pulsed with 25 µL of CD3/CD28-stimulating Dynabeads (Invitrogen Dynal AS, Oslo, Norway) in the presence of 30 U/mL of recombinant interleukin-2. Restimulation with the same concentrations was performed after 7 to 9 days, and subsequent restimulations were applied if deemed necessary based on cell numbers determined by microscopy and flow cytometry. After magnetic separation of the CD3+ T-cell fraction with MACS CD3 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's recommendation, CD3+ cell purity was routinely determined >96% by flow cytometry; in case of lower purity levels, a second purification was performed.
Preprocessing of gene expression and DMP
Two high-throughput data sets were used in this study: GEP and DMP data for 344 samples. GEP data were generated using Affymetrix HGU133 plus2.0 (Santa Clara, CA).13,14,25 Sample processing and quality control were carried out as described previously.13 Normalization of raw data were processed with Robust Multi-array Average,26,27 and probes on the array are remapped to RefSeq transcripts using a custom chip definition file (CDF)28 . The custom CDF mapped the original probes to known gene transcripts for University of California Santa Cruz HG19. DMP data were generated using the HpaII tiny fragment enrichment by ligation-mediated polymerase chain reaction assay, preprocessed as described previously,14 and annotated using University of California Santa Cruz HG19.
Preprocessing and detecting mutations in whole-exome sequence data
RAW-FASTQ files were aligned using Burrows-Wheeler Aligner29 followed by indel realignment using Genome Analysis ToolKit (GATK). The resulting aligned files (eg, BAM file) were then used to remove polymerase chain reaction duplicates using Sequence Alignment/Map tools.30 Single nucleotide variants were called using the unified genotyper of GATK, whereas all variants were annotated using Annovar. These annotations were subsequently used to select for nonsynonymous substitutions, stop-gain mutations, frameshift insertion, or frameshift deletions in the exonic or UTR5 regions that were not reported as a single nucleotide polymorphism, ie, by using the Single Nucleotide Polymorphism Database and the Cosmic database. Single nucleotide variants were also excluded if they were seen in the background, generated by whole exome sequencing of T cells of the same patient samples. Coverage and GATK statistics can be found in supplemental Table 2. The frequency of read depth of the aligned loci is illustrated in supplemental Figure 1.
Statistical analyses
Differentially expressed and methylated genes for the detected clusters are determined by comparing GEP and DMP data of each patient sample within the cluster vs patients outside the cluster, using the Student t test. Genes are considered to be differentially expressed or methylated when mRNA or DNA methylation levels differed with P ≤ .001 after correcting for multiple testing using the Benjamini and Hochberg31 method (denoted as the false discovery rate). Patient characteristics were compared using the Mann-Whitney U test (continuous variables) and the Fisher exact test (categorical variables) for 2-group comparisons, and Kruskal-Wallis test for 3-group comparison (continuous variables). Outcome measures are assessed using Kaplan-Meier estimates in a univariate analysis. Multivariate analyses were used according the Cox’s proportional hazard ratio model. The definition of complete remission (CR) and survival end points such as overall survival (OS), event-free survival (EFS), and relapse-free survival (RFS) were based on the recommended consensus criteria.32 Pathway analysis is performed by using the Molecular Signature Database, version 3.0, for the detection of enriched BioCarta pathways, Kyoto Encyclopedia of Genes and Genomes pathways, and transcription factor targets. Pathways and/or gene sets are considered statistically significant when the P value, derived from the hypergeometric test, is less or equal than .05 after correcting for multiple testing using the false discovery rate. In addition, pathways are derived using Ingenuity Pathway Analysis (Ingenuity Systems, http://www.ingenuity.com, IPA 8.8), with P ≤ .05.
Results
Hotspot mutations in SF genes SF3B1, U2AF35, and SRSF2 are more frequent in RAEB and AML-LBC than in AMLs
SF mutations have been reported to be present in MDS as well in AML. We screened complementary DNA of 47 RAEB, 29 AML-LBC, and 325 other AML patient samples (by dHPLC for 8 reported hotspot mutations)1 ie, in SF3B1 (5 hotspots: R625L/C; N626D; H662Q/D; K666N/T/E/R; K700E), U2AF35 (2 hotspots: S34F; Q157P), and SRSF2 (P95H/L/R). Samples with abnormal patterns were next confirmed by nucleotide sequencing. In 8 of 47 RAEB cases, we observed mutations in SRSF2. In 2 of 47 RAEB patients, mutations in SF3B1 were present. Twelve of the 29 AML-LBC patients carried SF mutations: U2AF35 (n = 4), SF3B1 (n = 1), and SRSF2 (n = 7) (Table 1). In the other AML samples, we found mutations in SF3B1 (n = 6), U2AF35 (n = 4), and SRSF2 (n = 15) (Table 2). We detected that the frequency of SF mutations in RAEB compared with AMLs is significant higher in the RAEB group (21.3% vs 7.69%, P < .001) and the AML-LBC group when compared with AMLs (41.4% vs 7.69%, P < .001). We detected no significant differences between frequencies of SF mutations in RAEB vs AML-LBC samples (Table 1). The latter 2 groups did not show significant differences in clinical characteristics, except for white blood cell (WBC) counts (5 × 109/L vs 14 × 109/L, P < .001), and BM blast percentages (11% vs 25%, P < .001, Table 1), with the latter being the parameter that a priori discriminates RAEB from AML.
We next compared the patients with SF mutations among RAEB, AML-LBC, and AMLs and detected that these 3 groups are highly similar in their clinical characteristics (Table 2). Again, differences were found in the expected BM blast percentages (P < .0001), and in WBC counts (P = .033). Thus, RAEB, AML-LBC, and AML cases with SF mutations share highly similar phenotypes, suggesting that these malignancies should be considered as 1 typical SF-mutant leukemic subset. This latter conclusion is further supported by our finding that AML patients with SF3B1, U2AF35, or SRSF2 mutations showed significant lower BM blast percentages than cases without SF mutations (51% vs 70%, P < .015; supplemental Table 3). Moreover, SF-mutant AML cases were older (58 vs 46 years, P < .0001), showed significantly lower WBC counts (25 × 109/L vs 36 × 109/L, P = .049) and had higher erythroblasts percentages (11% vs 3%, P < .0001; supplemental Table 3) than cases without SF mutations.
Two distinct RAEB/AML-LBC and AML-enriched clusters uncovered using integrative analysis of gene expression and cytosine methylation profiles
Gene expression and DNA-methylation data were available in a cohort of 9 RAEBs, 10 AML-LBCs, and 325 AMLs. We evaluated whether SF-mutant malignancies among these 344 patients carried unique GEP and DMP. We carried out 440 distinct hierarchical clustering analyses using different combinations of differentially expressed or differentially cytosine-methylated genes (supplemental Figure 2A). For each clustering, we addressed whether the grouping of samples was “stable” by computing the significance of the clusters with 1000 multiscale bootstraps. Subsequently, we computed the silhouette scores33 from the significant clusters, which describes how distinctive 1 cluster is from another. Using these statistics, we could computationally select the optimal hierarchical clustering. The selection of the most optimal combination of probe sets for clustering is explained in the “Computing the optimal hierarchical clustering” in the supplemental data. The optimal integrated hierarchical clustering was observed when GEP and DMP were combined using 2168 GEP and 2045 DMP probe sets, which resulted in the segregation of 18 clusters (Figure 1; supplemental Figure 2B). For each of the clusters, we assessed the enrichment for the currently known molecular and (cyto)genetical abnormalities. AMLs with either inv(16), t(15;17), or t(8;21) formed 3 distinct clusters each (clusters 1, 9, 10). CEBPA double-mutant and CEBPAsilenced AMLs formed clusters 16 and 18, respectively. Various other abnormalities (ie, mutations in NMP1, DNMT3A, IDH1 or IDH2, FLT3ITD, and FLT3TKD as well as chromosomal abnormalities 3q, 7q, or 11q23 defects) are depicted in Figure 1. The distribution of these well-characterized AML subsets using solely GEP or DMP data sets is represented in supplemental Figure 3. Detailed molecular and cytogenetic data of all AML patients in each cluster are presented in supplemental Table 4.
Hierarchical clustering of genetic and epigenetic features segregates AML patients into 18 clusters. Heat map representing pairwise correlations between the RAEB/AML patients using the gene expression and DNA-methylation profiles of each patient. Ordering of patient samples is based on hierarchical clustering using Pearson correlation and Ward’s linkage, which results in clusters of patients that are highly correlated to each other. Colored cells in the heat map depict a higher positive (red) or lower negative (blue) correlation, as indicated by the scale bar. Bars in the first 3 rows along the diagonal of the heat map indicate presence of the SF gene hotspot mutations. The last row indicates whether a patient is labeled as RAEB or AML-LBC. AML-LBCs are AML patients with blast counts between 20% and 30%. Detailed information of each patient in the clusters is shown in supplemental Table 4.
Hierarchical clustering of genetic and epigenetic features segregates AML patients into 18 clusters. Heat map representing pairwise correlations between the RAEB/AML patients using the gene expression and DNA-methylation profiles of each patient. Ordering of patient samples is based on hierarchical clustering using Pearson correlation and Ward’s linkage, which results in clusters of patients that are highly correlated to each other. Colored cells in the heat map depict a higher positive (red) or lower negative (blue) correlation, as indicated by the scale bar. Bars in the first 3 rows along the diagonal of the heat map indicate presence of the SF gene hotspot mutations. The last row indicates whether a patient is labeled as RAEB or AML-LBC. AML-LBCs are AML patients with blast counts between 20% and 30%. Detailed information of each patient in the clusters is shown in supplemental Table 4.
Besides the previously identified AML subgroups, 2 novel clusters [ie, 3 (n = 25) and 11 (n = 19)] were apparent. Clusters 3 and 11 are highly enriched for RAEB and AML-LBC patients (both P <.0001, Table 3). The unique GEP/DMP signatures that identified clusters 3 and 11 prompted further study.
Patients in clusters 3 and 11 are enriched for RAEB and AML-LBC with SF gene mutations
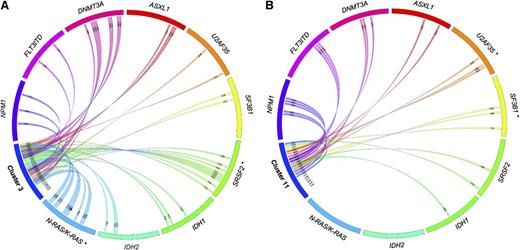
Of the 25 cases in GEP/DMP cluster 3, 4 are RAEB (16%; P < .0001, Table 3) and 5 are AML-LBC patients (20%; P < .0001, Table 3). The cluster was preferentially enriched for SF gene hotspot mutations (52%; 13/25, P < .0001), ie, SF3B1 (n = 2), U2AF35 (n = 2), and SRSF2 (n = 9) (Figures 1 and 2A; Table 3). SF mutations were detected in 2 of 4 RAEB, 4 of 5 AML-LBCs, and 7 of 16 AML cases (supplemental Table 5). The patients in cluster 11 were enriched for RAEB (21.1%, 4/19, P < .0001) and AML-LBC (26.3%, 5/19, P < .0001). Hotspot SF mutations were found in 8/19 (42.1%, P < .0001) of cases. The hotspot mutations are detected among SRSF2 (n = 2), SF3B1 (n = 3), and U2AF35 (n = 3) (Figures 1 and 2B; Table 3). SF mutations were seen in 3 of the 5 AML-LBC cases and 5 of the 10 AML cases (supplemental Table 5). When using the GEP or DMP data sets separately, no clusters were significantly enriched for patients with SF mutations (supplemental Figure 3). Thus the grouping of SF mutations was only evident when GEP and DMP data were used in combination.
Gene mutations in patients from clusters 3 and 11. Associations of gene mutations outlined by a Circos diagram for patients in cluster 3 (A) and cluster 11 (B). *Gene mutations that are significantly overrepresented for the particular cluster. SF mutations outside these clusters can be seen in supplemental Figure 5.
Gene mutations in patients from clusters 3 and 11. Associations of gene mutations outlined by a Circos diagram for patients in cluster 3 (A) and cluster 11 (B). *Gene mutations that are significantly overrepresented for the particular cluster. SF mutations outside these clusters can be seen in supplemental Figure 5.
We considered the possibility that in cases of clusters 3 and 11 that did not carry hotspot SF mutations that other SF alterations might be present. WES was carried out on DNA obtained from non–SF-mutant patients of which material was available (ie, 7 samples from cluster 3 and 7 from cluster 11). We did not find other mutations in any of the 8 SF genes previously reported to be frequently mutated. However, 3 acquired mutations (absent in T cells from the same patients) were found in other RNA-binding or RNA-SF genes. In cluster 11, mutations in DHX15 (nonsynonymous; patient 6448), PRPF4B (frameshift deletion; patient 2246), and CELF4 (nonsynonymous; patient 3318) were found (supplemental Table 6).
Erythroid phenotype of cluster 11 patient samples
Morphological analysis of BM samples from patients in clusters 3 and 11 revealed that blast percentages of the 2 clusters were both significantly lower compared with the patients outside cluster 3 (34% vs 68%, P < .0001) and cluster 11 (31% vs 68%, P < .0001, Table 3). Higher percentages of erythroblasts were found in cluster 11 marrow preparations when compared with the other AMLs (32% vs 3%, P < .0001, Table 3). WBC counts of cluster 11 cases were significantly reduced in comparison with unselected AMLs (P < .0001; 6 × 109/L; vs 36 × 109/L, respectively), whereas cluster 3 patients showed WBC counts that were equal to patients outside cluster 3 (31 × 109/L, Table 3). Thus the 2 SF-mutant clusters show morphological differences for which cluster 11 patients revealed a strong erythroid phenotype.
Differentially expressed or hypomethylated genes in cluster 11 patient samples strongly associate with erythroid development
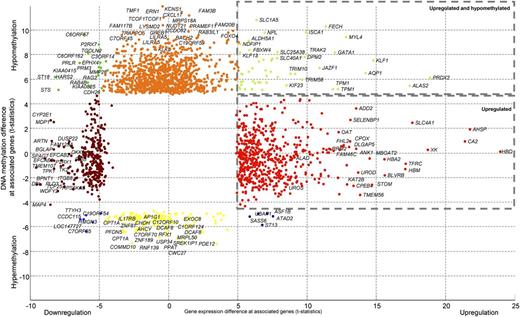
The signature of 895 differentially expressed and 1180 differentially methylated genes characterized the cases in cluster 11 compared with unselected AMLs (supplemental Table 7). Pathway analysis revealed that the profiles in cluster 11 were highly enriched for gene sets associated with erythroid development and function, eg, α-hemoglobin stabilizing protein pathway,34,35 porphyrin metabolism, or P53 signaling (Figure 3; supplemental Table 8). Numerous erythroid genes were found to be hypomethylated and comparatively overexpressed, such as GATA1, FECH, ALAS2, AQP1, or KLF1. Other erythroid genes were overexpressed with no change in DNA-methylation, such as for ALAD, UROS, UROD, AHSP, or HBD (supplemental Figures S4 and S5). Analysis of transcription factor–binding sites using the differentially expressed and methylated genes revealed significant enrichment for the E2F and GATA1 transcription factor–binding sites among these genes (P < .002 and P < .001, respectively; supplemental Table 8).
Specific DNA-methylation and gene expression patterns for patient samples from cluster 11. Differential expressed and DNA-methylation genes in patient samples from cluster 11 compared with all other AMLs are indicated with different colored dots. The colors depict the gene expression and DNA-methylation status, ie, the right upper corner represents genes that are hypomethylated and overexpressed (green dots). Many of these genes encode for proteins involved in erythroid development or function (detailed results are depicted in supplemental Table 7).
Specific DNA-methylation and gene expression patterns for patient samples from cluster 11. Differential expressed and DNA-methylation genes in patient samples from cluster 11 compared with all other AMLs are indicated with different colored dots. The colors depict the gene expression and DNA-methylation status, ie, the right upper corner represents genes that are hypomethylated and overexpressed (green dots). Many of these genes encode for proteins involved in erythroid development or function (detailed results are depicted in supplemental Table 7).
In contrast to the cluster 11 patient group, the 1522 differentially expressed and 74 methylated genes that are associated with cluster 3 lacked the dominant erythroid signature (supplemental Figure 6A,B). Although expression of some erythroid genes is seen, these were not significantly higher compared with other patients. Thus although both clusters 3 and 11 are enriched for RAEB and AML-LBC cases and frequently harbor SF mutations, cluster 11 cases are specifically associated with a combined myeloid and erythroid phenotype.
Cluster 3 patient group frequently carries RAS mutations and has unfavorable outcomes
Whole-exome sequencing on the small selection of cluster 3 and 11 cases revealed 1 KRAS and 3 NRAS mutants among the 6 cases of cluster 3 that were analyzed. We applied Sanger sequencing for NRAS and KRAS among all patients of the 2 clusters. Ten of the 25 (40%) patients in cluster 3 carried mutations in NRAS (n = 9) or KRAS (n = 1) (P < .0001; Table 3 and Figure 2). In contrast, no RAS mutations were found in any of the cluster 11 cases analyzed (Table 3).
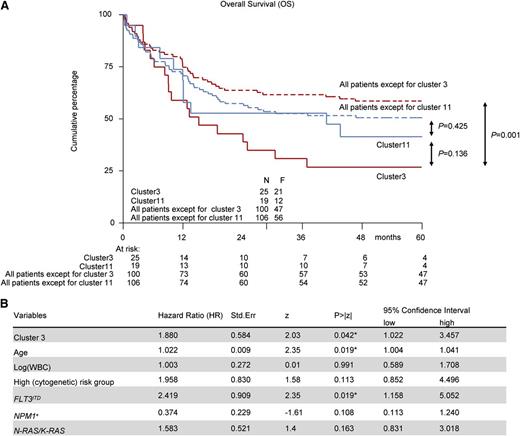
To verify whether cluster 3 and 11 differed clinically in terms of prognosis, we assessed the OS, RFS, and EFS. The OS for patients in cluster 3 and 11 showed a 5-year OS of 24% (95% confidence interval, 9-42) and 41% (95% confidence interval, 20-62) respectively (Figure 4). In a univariate analysis, cluster 3 patients showed significantly inferior outcome measures compared with unselected AMLs (OS: P = .001, Figure 4A; RFS: P = .014, supplemental Figure 7A; EFS: P = .016, supplemental Figure 7B), whereas this was not seen for cluster11 cases (OS: P = .425, Figure 4A; RFS: P = .944, supplemental Figure 7A; EFS: P = .638, supplemental Figure 7B). In a multivariate analysis, we could confirm that cluster 3 patients showed a poor treatment response, independent from other relevant covariates with prognostic value (age, WBC count, FLT3ITD, NPM1pos, NRAS/KRAS, and high-risk [cyto]genetics) (OS: P = .042; Figure 4B; RFS: P = .045; supplemental Figure 7C; EFS: P = .1; supplemental Figure 7D). The multivariate analysis did not reach significance for cluster 11 (supplemental Figure 7E,F).
Survival analysis for patients in clusters 3 and 11. Kaplan-Meier survival curves and multivariate analysis for OS. Multivariate analysis is based on the Cox proportional hazard ratio (HR) model. The included variables in the model are: NPM1mut vs wild-type NPM1, FLT3ITD vs no FLT3ITD, NRASmut/KRASmut vs wild-type NRAS/KRAS, and high cytogenetic risk vs no high cytogenetic risk. Age and WBC count are used as a continuous variable. (A) Kaplan-Meier curves for cluster 3 vs all patients except cluster 3 patients, cluster 11 vs all patients except cluster 11 patients, and cluster 3 vs cluster 11 patients. (B) Multivariate analysis for cluster 3 patients.
Survival analysis for patients in clusters 3 and 11. Kaplan-Meier survival curves and multivariate analysis for OS. Multivariate analysis is based on the Cox proportional hazard ratio (HR) model. The included variables in the model are: NPM1mut vs wild-type NPM1, FLT3ITD vs no FLT3ITD, NRASmut/KRASmut vs wild-type NRAS/KRAS, and high cytogenetic risk vs no high cytogenetic risk. Age and WBC count are used as a continuous variable. (A) Kaplan-Meier curves for cluster 3 vs all patients except cluster 3 patients, cluster 11 vs all patients except cluster 11 patients, and cluster 3 vs cluster 11 patients. (B) Multivariate analysis for cluster 3 patients.
Discussion
In this study, we evaluated the frequency of SF mutations in RAEB, AML-LBC, and AML patients. We demonstrate that the decision to define RAEB as MDS and AML-LBC as AML, solely based on percentage of blasts, is artificial and that SF-mutant AML/RAEB should be viewed as a related disease entity. Second, we studied GEP and DMP data in a considerable cohort of patients with AML and RAEB with a particular focus on patients with SF mutations. Based on combined GMP/DMP-data, 2 distinct SF-mutant RAEB/AML subtypes could be recognized, which differ morphologically, molecularly, and clinically from each other. Not all patients in the 2 clusters that we identified carried 1 of the currently well-described hotspot mutations in the SF genes SF3B1, U2AF35, and SRSF2. Because we focused in this study on hotspot mutations, we did not analyze the presence of the previously reported mutations in ZRSR2, SF1, or PRPF40B. It is possible that deep-sequencing procedures including these genes in the analysis may lend further support to the conclusions that we draw in this study, and that such future studies will be of interest. Moreover, it is also possible that, using massive parallel deep sequencing, yet another cluster may be uncovered that has been missed by focusing on hotspot mutations in SF3B1, U2AF35, and SRSF2.36 Nevertheless, the current study provides novel data that point to the existence of mutations in other genes encoding RNA-binding/splicing factors. Although our detected mutations in DHX15, PRPF4B, and CELF4 have not previously been reported in AML, other DHX and PRPF family members have been found in AML and MDS as reported in The Cancer Genome Atlas (http://cancergenome.nih.gov/). Mutations in DHX15, PRPF4B, and CELF4 have been reported in the COSMIC database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/). Moreover, Yoshida et al1 reported mutations in the SF gene PRPF40B. Together, these observations favor the hypothesis that more SF genes may be mutated in AML/RAEB/AML-LBC patients that can be uncovered using GEP and DMP data sets in a combined manner.
It has previously been demonstrated that distinct molecularly defined AML subtypes cluster using gene expression or DNA-methylation profiles in an unsupervised manner.13,14 In contrast to AMLs with, for instance, translocations t(8;21) and t(15;17) or with mutations in CEBPA, SF-mutant malignancies did not form any unique cluster when GEP or DMP data were used separately. It was only through integrating these data sets that we were able to identify SF-mutant patients as being distinct from other cases and consisting of 2 subgroups. These clusters can only be discovered by combining data sets because the gene expression profiles or DNA-methylation profiles separately do not yield sufficient discriminative features to distinguish these patients according the 2 identifiable clusters. Cluster 11 contains patients with strongly expressed as well as hypomethylated erythroid genes, as illustrated in Figure 3. Increased mRNA expression levels of erythroid genes are also seen in patients of other clusters, but not in combination with hypomethylation. These combined features, among others, could explain why these patients clustered so strongly when GEP data and DMP data were used in an integrated manner (Figure 1). Why patients from cluster 3 could only be defined using the combination of GEP and DMP data sets is not as clear as for cluster 11. However, based on the silhouette scores using the bootstrap labels from Pvclust (see the supplemental material), the hierarchical clustering appeared stable. That samples in this group were enriched for SF mutations and RAS mutations emphasizes that with combined GEP/DMP data sets, a unique signature could be derived that recognizes a leukemia subgroup.
We found multiple SF-mutant samples outside clusters 3 and 11. The question is whether these SF-mutant AMLs are biologically different or whether they were grouped in different clusters because of technical inaccuracies, meaning that they should have been identified as cluster 3 or 11 cases when more sophisticated procedures of gene expression and genomewide cytosine methylation analyses had been applied. Gene chip hybridization experiments that we applied in this study is now being replaced by RNA-Seq, a procedure that not only determines gene expression levels, but also discriminates between different splice forms. To study cytosine methylation, we applied the HpaII tiny fragment enrichment by ligation-mediated polymerase chain reaction, an assay that generates “snapshots” of small areas within CpG-rich regions. We hypothesize that the combination of RNA-Seq with more sophisticated tools to determine DNA-methylation profiles will provide information that will allow us to generate even better combined GEP/DMP signatures. It is possible that SF-mutant cases that were not found in clusters 3 or 11 potentially belong to either of these 2 clusters but were missed with the currently used methodologies. In any case, our study highlights the potential of combining biological data sets such as gene expression and DNA-methylation profiling data and shows that with pursuing such a combined approach, leukemia subtypes with a characteristic genotype hidden among the heterogeneity can be uncovered.
Novel cluster 11 was most remarkable for involving MDS and AML because these samples appeared to share unique erythroid features based on the following findings: (1) Enrichment of pathways associated with erythroid development, when differentially expressed and methylated genes were analyzed; (2) multiple erythroid genes were simultaneously highly expressed and hypomethylated; (3) high cytological percentages of erythroblasts; (4) presence of patient samples with RAEB or AML-LBC; and (5) a frequent appearance of ring sideroblasts. These AMLs showed differential expression and hypomethylation of erythroid genes as well. We conclude that AMLs with defective erythroid development exist more frequently than morphological classification would suggest.
The 2 RAEB/AML clusters show several differences, among which are the high percentages of N-RAS or K-RAS mutations in cluster 3 but not cluster 11 patients. This striking difference between the SF-mutant–enriched clusters may explain the much higher WBC counts found among cluster 3 samples. Cluster 3 patients also contain more frequent mutations in SRSF2, which has been reported to occur in AMLs that develop upon leukemic transformation from myeloproliferative neoplasms. We hypothesize that the 2 clusters we identified represent 2 different SF mutant malignancies, which may embody distinct evolutionary stages of the disease. This would mean that certain cases in cluster 11 may become cluster 3 AMLs in a later phase of the disease (ie, upon acquiring mutations in N-RAS or K-RAS). No matter the explanation, our data strongly suggest that SF-mutant RAEB and AML constitute a myeloid entity that overrides the separation between AML and MDS and is composed of 2 subgroups that show overlap but also differ clinically and molecularly.
There is an Inside Blood Commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank their colleagues at the Bone Marrow Transplantation Group and the Molecular Diagnostics Laboratory of the Department of Hematology at Erasmus University Medical Center (Erasmus MC) for storage of samples and molecular analysis of leukemia cells.
This work was supported by a grant from ErasmusMC (MRace) (R.D., E.T.) and a research fellowship from the Dutch Cancer Society “Koningin Wilhelmina Fonds.” This research was performed within the framework of the Center for Translational Molecular Medicine, project BioCHIP (grant 03O-102).
Authorship
Contribution: E.T., R.D., and B.L. conceived and designed the study; P.J.M.V., B.L., A.M., and R.D. provided study materials or patients; E.T., M.H., K.v.L., M.A.S., E.B., Y.v.N., R.H., M.J.T.R., M.E.F., P.J.M.V., B.L., A.M., and R.D. collected and assembled data; E.T., B.L., A.M., and R.D. analyzed and interpreted data; E.T., R.D., and B.L. wrote the manuscript; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ruud Delwel, Erasmus University Medical Center, Department of Hematology, PO Box 2040, 3000CA Rotterdam, The Netherlands; e-mail: h.delwel@erasmusmc.nl.