Abstract
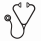
INTRODUCTION: Patients with lower-risk myelodysplastic syndromes (MDS) as defined by the International Prognostic Scoring System (IPSS) are generally considered to have favorable prognosis. The MD Anderson Lower-risk Prognostic Scoring system (MDA LR-PSS) is a validated prognostic model that defines a subset of patients with shorter than expected survival. Presence of EZH2mutations has been reported to independently impact survival of these patients. Further evaluation and validation of the addition of mutation data to this prognostic model is needed.
METHODS: We conducted whole exome sequencing of 74 previously untreated patients with Low or Int-1 IPSS. Exome capture hybrid was performed using Agilent SureSelect All Exon V4. Sequencing was performed with Illumina HiSeq 2000 and aligned to the hg19 human genome reference. Common virtual normal in house pool was used for somatic variant calling. Generalized linear models were used to study association of overall response (OR), complete response (CR) and risk factors. Response was defined following 2006 IWG criteria. The Kaplan-Meier produce limit method was used to estimate the median overall survival (OS) and leukemia-free survival (LFS).
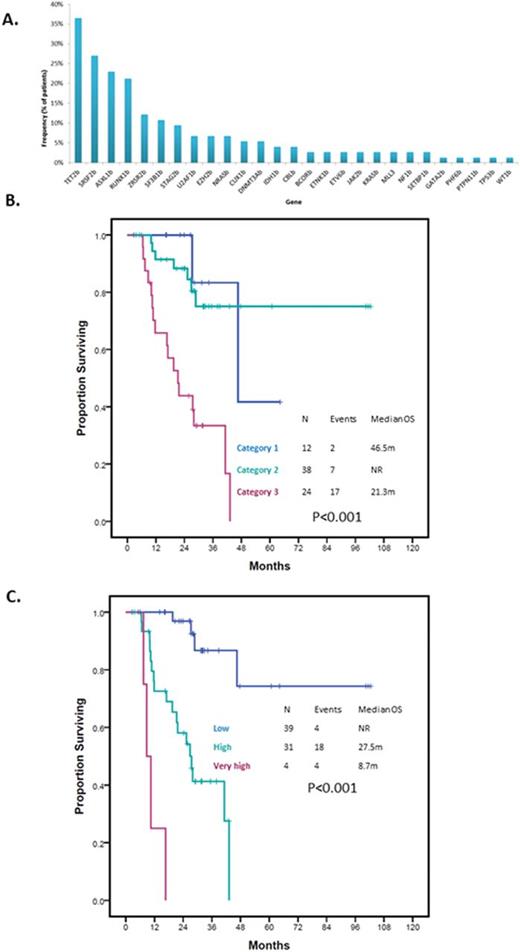
RESULTS: Patient characteristics are summarized in Table 1. Seventeen (23%) patients had CMML and 57 (77%) had MDS. Seventy (95%) patients had evaluable cytogenetics. A total of 46 (66%) patients had normal karyotype with 4 (6%) patients having complex karyotype. Twenty-three (31%) patients were classified as Low risk by IPSS and 51 (69%) as Int-1. Twelve (16%) patients were classified as Category 1 by the MDA LR-PSS, 38 (51%) as Category 2 and 24 (32%) as Category 3. A total of 148 driver mutations in 27 genes were detected. Median number of driver mutations per patient was 2 (range 0-5). Mutations in TET2, SRSF2, ASXL1, SF3B1 and ZRSR2 were the most frequently detected mutations present in >10% cases. Frequency of identified mutations is shown in Figure 1A. The median follow up was 26.5 months (range 3-102 months). Median OS was 43.2months (95% CI 35.04-50.46). Survival by MDA LR-PSS category is shown in Figure 1B. A total of 6 patients had transformation to acute myeloid leukemia with a median time to transformation of 22.4 months (95% CI 0.00-45.28). By univariate analysis, mutations in BCOR (HR 5.49, 9% CI 1.24-24.29, p=0.011), RUNX1 (HR 2.8, 95% CI 1.11-7.09, p=0.023) and STAG2 (HR 3.32, 95% CI 1.12-9.82, p=0.022) predicted for shorter OS. When analyzing for LFS, mutations in EZH2 (HR 18.61, 95% CI 2.99-115.92, p<0.001) and U2AF1 (HR 5.08, 95% CI 0.92-28.06, p=0.038) predicted for shorter LFS. By multivariate analysis combining the identified driver mutations with prognostic relevance and the MDA LR-PSS category, presence of MDA LR-PSS Category 3 (HR 5.98, 95%CI 1.28-28, p=0.023), RUNX1 mutation (HR 4.18, 95%CI 1.36-12.81, p=0.012) and STAG2 mutation (HR 3.7, 95%CI 1.02-13.47, p=0.047) retained there prognostic significance. Based on these results we designed a new model with scoring being based on HR for each given variable: MDA LR-PSS Category 3 2 points, RUNX1mut 1 point and STAG2mut 1 point. Patients were classified into three categories: Low, High or Very high with significantly different survival outcomes (Figure 1C). Based on this new model, patients with Category 2 MDA LR-PSS with either STAG2 or RUNX1 mutations had similar outcomes to those with Category 3 and no mutations (p=0.747). Patients with either Category 1 or Category 2 by MDA LR-PSS without STAG2 or RUNX1 mutations had similar OS (p=0.306) and represented the population with most favorable outcomes (p<0.001). Finally, patients with Category 3 with STAG2 or RUNX1 mutations had the worse outcomes in terms of survival (p<0.001).
CONCLUSION: Similar to previous studies, our data suggests integration of mutation data into the MDA LR-PSS can improve our ability to predict outcomes of patients with lower-risk MDS. Mutations in STAG2 and RUNX1 can help identify a subset of patients with worse than expected outcomes as predicted by the MDA LR-PSS.
Jabbour:ARIAD: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; Novartis: Research Funding; BMS: Consultancy. Ravandi:Seattle Genetics: Consultancy, Honoraria, Research Funding; BMS: Research Funding. Cortes:ARIAD: Consultancy, Research Funding; BMS: Consultancy, Research Funding; Novartis: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; Teva: Research Funding. DiNardo:Daiichi Sankyo: Other: advisory board, Research Funding; Abbvie: Research Funding; Novartis: Other: advisory board, Research Funding; Celgene: Research Funding; Agios: Other: advisory board, Research Funding. Daver:Otsuka: Consultancy, Honoraria; Kiromic: Research Funding; Ariad: Research Funding; BMS: Research Funding; Sunesis: Consultancy, Research Funding; Karyopharm: Honoraria, Research Funding; Pfizer: Consultancy, Research Funding. Konopleva:Reata Pharmaceuticals: Equity Ownership; Abbvie: Consultancy, Research Funding; Genentech: Consultancy, Research Funding; Stemline: Consultancy, Research Funding; Eli Lilly: Research Funding; Cellectis: Research Funding; Calithera: Research Funding. Kantarjian:Bristol-Myers Squibb: Research Funding; ARIAD: Research Funding; Amgen: Research Funding; Pfizer Inc: Research Funding; Delta-Fly Pharma: Research Funding; Novartis: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract