Key Points
CDK6 acts as a transcriptional regulator to suppress Egr1 in HSCs and LSCs, allowing their activation.
Cdk6−/− HSCs fail to contribute to repopulation in competitive transplants, and BCR-ABLp210+Cdk6−/− LSCs fail to inflict disease.
Abstract
The cyclin-dependent kinase 6 (CDK6) and CDK4 have redundant functions in regulating cell-cycle progression. We describe a novel role for CDK6 in hematopoietic and leukemic stem cells (hematopoietic stem cells [HSCs] and leukemic stem cells [LSCs]) that exceeds its function as a cell-cycle regulator. Although hematopoiesis appears normal under steady-state conditions, Cdk6−/− HSCs do not efficiently repopulate upon competitive transplantation, and Cdk6-deficient mice are significantly more susceptible to 5-fluorouracil treatment. We find that activation of HSCs requires CDK6, which interferes with the transcription of key regulators, including Egr1. Transcriptional profiling of HSCs is consistent with the central role of Egr1. The impaired repopulation capacity extends to BCR-ABLp210+ LSCs. Transplantation with BCR-ABLp210+–infected bone marrow from Cdk6−/− mice fails to induce disease, although recipient mice do harbor LSCs. Egr1 knock-down in Cdk6−/− BCR-ABLp210+ LSKs significantly enhances the potential to form colonies, underlining the importance of the CDK6-Egr1 axis. Our findings define CDK6 as an important regulator of stem cell activation and an essential component of a transcriptional complex that suppresses Egr1 in HSCs and LSCs.
Introduction
A cyclin-dependent kinase (CDK) is a critical regulator of cell-cycle progression, becoming activated upon binding to cyclins. Progression through the G1 phase of the cell cycle is mediated by activation of the CDK4/6-cyclinD complex and subsequent phosphorylation of the retinoblastoma protein, which triggers E2F-dependent transcription.1,2 CDK4 and CDK6 show 71% amino acid homology and have been considered to fulfill largely redundant functions because only the simultaneous deletion of both genes leads to embryonic lethality resulting from hematopoietic defects.3,4 Cdk6 deficiency is characterized by subtle defects in the hematopoietic system, such as defects in thymocyte development and a reduction in erythrocyte numbers.4,5 CDK6 has been shown to have a kinase-independent function in myeloid cells, where it interacts with RUNX1 to block RUNX1-dependent transcription.6 We recently discovered a key role for CDK6 in lymphoma formation: CDK6 transcriptionally regulates Vegf-A and p16INK4a by interacting with signal transducer and activator of transcription (STAT) and AP-1 transcription factors.7 A subsequent report described CDK6 as a transcriptional coregulator of nuclear factor κB p65.8 CDK6 appears to have a key role in hematopoietic tumors, where it is frequently upregulated.5,7 CDK6 has also been shown to be critical in acute myeloid leukemia (AML) and acute lymphoblastic leukemia driven by mixed lineage leukemia fusion proteins.9,10 There is considerable interest in targeting CDK4/6 in cancer therapy, and the Food and Drug Administration nominated CDK4/6 inhibitors as the “breakthrough therapeutic advance” in 2013.
All hematopoietic cells arise from hematopoietic stem cells (HSCs), which possess the ability to self-renew and to differentiate into all blood cell lineages.11 The existence of a deeply dormant HSC (d-HSC) population that only divides 5 times was recently postulated.12 d-HSCs are activated in response to injury signals such as 5-fluorouracil (5-FU) treatment but are not thought to have any function in steady-state homeostasis.12 Under stress conditions, d-HSCs enter the cell cycle, leading to a rapid increase in numbers of multipotent progenitors (MPP), which differentiate into distinct mature hematopoietic cells. The activation of quiescent d-HSCs is controlled by a network of transcriptional regulators such as EGR1, RUNX1, SCL, and PBX1.13-16
Our understanding of the molecular networks that control HSCs is currently fragmentary. Research on leukemia provided the first striking observations for the existence of cancer stem cells. In AML and chronic myeloid leukemia (CML), only a limited population of cells is able to maintain and transduce disease. These so-called leukemic stem cells (LSCs) frequently express surface markers indicative of HSCs, and HSCs and LSCs clearly share functional characteristics and gene expression profiles.17,18 LSCs and HSCs have unlimited capacity for self-renewal, and Bmi-1,19 Wnt/β-Catenin,20 and the Hedgehog pathway are activated.21 There is evidence that LSCs may remain dormant for a long period (up to10 years),22 explaining the high rate of relapse of leukemia patients after extended periods of remission. LSCs thus remain a major therapeutic challenge.
We describe a new facet of the regulation of HSCs and LSCs. Cdk6-deficient HSCs lack the ability to repopulate in competitive transplant experiments and fail to respond adequately to challenge with polyinosinic:polycytidylic acid (poly(I:C)) and 5-FU. Furthermore, Cdk6−/− BCR-ABLp210+ LSCs fail to repopulate upon transplantation. These results identify CDK6 as a crucial player in the activation of HSC and LSCs.
Methods
Mouse strains
All mice were maintained under pathogen-free conditions at the University of Veterinary Medicine, Vienna, Austria. Ly5.1+(CD45.1+), wild-type, and Cdk6−/− (from M. Malumbres4 ) mice were kept on a C57Bl/6J background. NOD/SCID/IL-2Rγ−/− (NSG) mice were obtained from The Jackson Laboratory (USA). Six- to 8-week-old mice were used for experiments unless indicated elsewise. Animal experiments were performed in accordance with protocols approved by the Austrian law and the Animal Welfare Committee at the Veterinary University of Vienna.
Cell-cycle analysis
Cell-cycle analysis was performed in a 2-step protocol by staining with antibodies directed against HSC surface markers followed by intracellular staining with Ki-67 and 4,6 diamidino-2-phenylindole (DAPI). For intracellular staining, cells were fixed in Cytofix/Cytoperm (BD Biosciences), washed and stained with Ki-67 FITC (BD Bioscience), and then costained with 0.1 mg/100 mL DAPI in phosphate-buffered saline (PBS) with 0.1% Triton X-100 (Sigma-Aldrich).
Transplantation studies
For competitive transplantation assays, the bone marrow (BM) of the Cdk6+/+(Ly5.1+) and Cdk6−/−(Ly5.2+) mice were mixed at a ratio of 1:9, 1:1 or 9:1 and injected intravenously into lethally irradiated (9 Gy) C57Bl/6J mice (in total 5 × 106 cells/mouse). The irradiated control mice died after 9 days. For the assessment of long-term repopulation capacities of transplanted HSCs, mice were euthanized 16 weeks posttransplantation. BM, spleens, and peripheral blood were analyzed by fluorescence-activated cell sorter (FACS) for the contribution of Ly5.1+ and Ly5.2+ cells to individual LSK populations and mature lymphoid (CD19+, CD3+) and myeloid lineages (Gr1+ Mac1+).
Transcriptional profiling
Total RNA was extracted from the FACS fraction A cells (Lin−Sca1+c-Kit+CD150+CD48−) using the RNeasy Micro Kit (Qiagen). The RNA samples were quality controlled using the Laboratory-Chip technique (Agilent Bioanalyzer) and subsequently preamplified according to the TransPlex Whole Transcriptome Amplification WTA2 protocol (Sigma-Aldrich). Samples were then fluorescently labeled by in vitro transcription using the Two-Color Microarray-Based Gene Expression Analysis kit (Agilent) and hybridized onto Mouse Gene Expression G3 60K arrays (Agilent) containing ∼56,000 60-mer probes. Images were acquired and quantified by confocal scanner and software (Agilent G2505C and Feature Extraction). Expression levels were processed using standard methods of normalization and significance analysis as described previously.23 A multiple testing correction with false discovery rate adjustment by the Benjamini-Hochberg method was performed. Gene ontology and pathways were analyzed using Ontologizer,24 JASPAR,25 and GeneMANIA databases.26 Heatmaps were generated using Caleydo software.27
Statistical analysis
Data are reported as mean values ± standard deviation and were analyzed by GraphPad. Differences were assessed for statistical significance by Student t test or 1-way analysis of variance. Kaplan-Meier plots were analyzed by the log-rank test. Statistical significance is as follows: *P < .05, **P < .01, ***P < .001, ****P < .0001.
Homing assay
Competitive setting.
Cdk6+/+ and Cdk6−/− BM cells were seeded on GP+E86 retroviral producer cells (pMSCV-IRES-GFP or pMSCV-IRES-dsRed) in Dulbecco’s modified Eagle medium containing 25 ng/mL IL-3, 50 ng/mL IL-6, 50 ng/ml stem cell factor (SCF), and 7 µg/mL polybrene. After 48 hours incubation, equal numbers (100 000 cells/mouse) of Cdk6+/+ dsRed+ LSKs and Cdk6−/− GFP+ LSKs were injected intravenously into lethally irradiated (9 Gy) Cdk6+/+Ly5.2+ animals together with 3 × 106 LSK–depleted BM carrier cells. After 18 hours, mice were euthanized and BMs were analyzed for the presence of dsRed+ and GFP+ LSKs.
Noncompetitive setting.
Cdk6+/+Ly5.2+ and Cdk6−/−Ly5.2+ BM was sorted by FACS, and 1 × 106 cells (containing comparable numbers of LSKs) were injected into lethally irradiated Cdk6+/+Ly5.1+ mice. After 18 hours, mice were euthanized and the BM was analyzed for the presence of Ly5.2+ LSKs by FACS.
Results
Cdk6−/− HSCs fail to repopulate efficiently in a competitive transplant setting
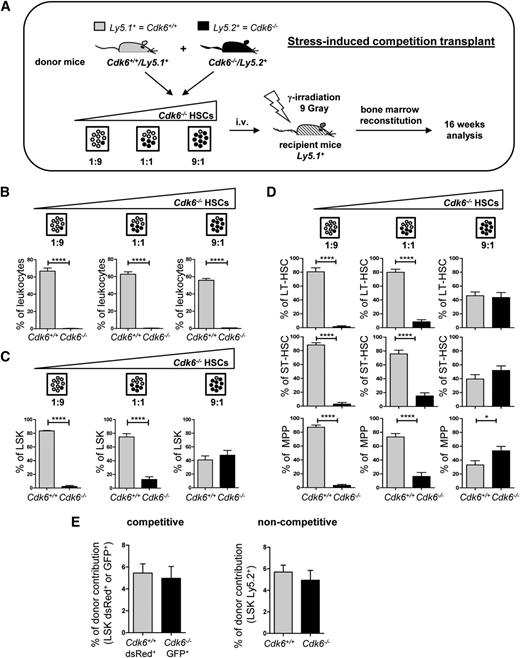
CDK6 is expressed at high levels in hematopoietic cells, including HSCs (http://biogps.org/). To investigate the role of CDK6 in HSCs, we set up competitive transplant experiments using Cdk6−/− mice. Distinct ratios of Cdk6+/+Ly5.1+ and Cdk6−/−Ly5.2+ BM cells were transplanted into lethally irradiated Ly5.1+ mice (Figure 1A). Sixteen weeks later, the contribution of Cdk6+/+Ly5.1+ and Cdk6−/−Ly5.2+ cells to the individual hematopoietic cell populations were analyzed (supplemental Figure 1A, available on the Blood Web site). Even when only a small proportion of donor cells were Cdk6+/+, the majority of the leukocytes in the BM were derived from them (Figure 1B). Analysis of individual hematopoietic lineages revealed no Cdk6−/−/Ly5.2+ Gr1+Mac1+ or CD19+ and only low numbers of CD3+ cells (which most likely represent long-lived T cells) (supplemental Figure 1B). The spleen and blood contained very low numbers of Cdk6−/−/Ly5.2+ cells (data not shown). Remarkably, there were significant numbers of Cdk6−/− LSK (Lin−Sca1+c-Kit+) cells in the BM, although they did not contribute to the pool of differentiated cells (Figure 1C). There were no detectable changes in the distribution of the LSK pool in recipient mice: the proportions of long-term HSCs, short-term HSCs, and MPP reflected the total LSK population (Figure 1D). Competitive (ratio 1:1) and noncompetitive homing experiments confirmed that the results could not be attributed to homing defects in Cdk6−/− LSKs (Figure 1E).
CDK6 is required for reconstitution of the hematopoietic system. (A) Schematic representation of experimental design. Cdk6+/+/Ly5.1+ and Cdk6−/−/Ly5.2+ BM cells were transplanted in ratios of 1:9, 1:1, and 9:1 into lethally irradiated (9 Gy) wild-type recipient mice. Long-term BM reconstitution was analyzed 16 weeks after transplantation (n = 5 per genotype; Cdk6+/+ and Cdk6−/− groups were compared using Student t test). (B) Ly5.1+/Ly5.2+ compositions were analyzed in total BM leukocytes (n = 5 per genotype; ****P < .0001). The leukocytic population included lymphocytes and myeloid cells, but excluded debris and erythrocytes, as determined by forward scatter/side scatter blots. (C) LSK cells were analyzed for Ly5.1+/Ly5.2+ composition in each transplantation setting (n = 5 per genotype; ****P < .0001). (D) Contributions of Ly5.1+ and Ly5.2+ cells in long-term HSCs (LT-HSCs), short-term HSCs (ST-HSCs), and MPP (n = 5 per genotype; ****P < .0001, *P < .05). (E) Cdk6+/+ and Cdk6−/− BM were infected with empty dsRed+ or GFP+ retrovirus. Equal numbers of Cdk6+/+ dsRed+ LSKs and Cdk6−/− GFP+ LSKs (100 000/mouse) were injected in a 1:1 ratio into lethally irradiated recipient animals in a competitive setting (LSK-depleted carrier BM was added and a total of 3 × 106 cells/mouse were injected). After 18 hours, mice BM was analyzed for dsRed+ and GFP+ LSKs (left panel, n = 6). In a noncompetitive setting, equal numbers of either Cdk6+/+/Ly5.2+ or Cdk6−/−/Ly5.2+ BM cells were injected into lethally irradiated Ly5.1+ mice (1 × 106 cells/mouse). After 18 hours, BM was analyzed for the presence of donor-derived Ly5.2+ LSKs (right panel, n = 4 per genotype).
CDK6 is required for reconstitution of the hematopoietic system. (A) Schematic representation of experimental design. Cdk6+/+/Ly5.1+ and Cdk6−/−/Ly5.2+ BM cells were transplanted in ratios of 1:9, 1:1, and 9:1 into lethally irradiated (9 Gy) wild-type recipient mice. Long-term BM reconstitution was analyzed 16 weeks after transplantation (n = 5 per genotype; Cdk6+/+ and Cdk6−/− groups were compared using Student t test). (B) Ly5.1+/Ly5.2+ compositions were analyzed in total BM leukocytes (n = 5 per genotype; ****P < .0001). The leukocytic population included lymphocytes and myeloid cells, but excluded debris and erythrocytes, as determined by forward scatter/side scatter blots. (C) LSK cells were analyzed for Ly5.1+/Ly5.2+ composition in each transplantation setting (n = 5 per genotype; ****P < .0001). (D) Contributions of Ly5.1+ and Ly5.2+ cells in long-term HSCs (LT-HSCs), short-term HSCs (ST-HSCs), and MPP (n = 5 per genotype; ****P < .0001, *P < .05). (E) Cdk6+/+ and Cdk6−/− BM were infected with empty dsRed+ or GFP+ retrovirus. Equal numbers of Cdk6+/+ dsRed+ LSKs and Cdk6−/− GFP+ LSKs (100 000/mouse) were injected in a 1:1 ratio into lethally irradiated recipient animals in a competitive setting (LSK-depleted carrier BM was added and a total of 3 × 106 cells/mouse were injected). After 18 hours, mice BM was analyzed for dsRed+ and GFP+ LSKs (left panel, n = 6). In a noncompetitive setting, equal numbers of either Cdk6+/+/Ly5.2+ or Cdk6−/−/Ly5.2+ BM cells were injected into lethally irradiated Ly5.1+ mice (1 × 106 cells/mouse). After 18 hours, BM was analyzed for the presence of donor-derived Ly5.2+ LSKs (right panel, n = 4 per genotype).
Cdk6−/− mice harbor increased levels of quiescent HSCs
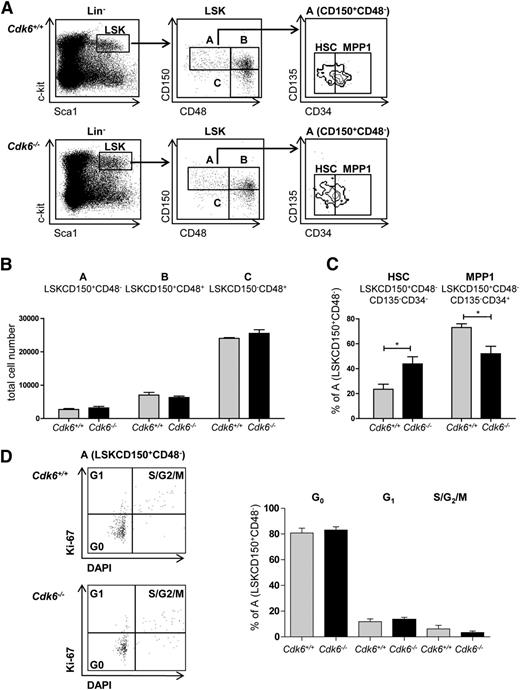
Under homeostatic conditions, the numbers of cells in the BM of Cdk6−/− animals was comparable to those of controls (supplemental Figure 2A)4 and the total LSK numbers were unaltered (supplemental Figure 2B). Examination of individual LSK subpopulations using SLAM markers12,28 (Figure 2A) failed to reveal significant alterations in the distribution of cells in fraction A (LSKCD150+CD48−), fraction B (LSKCD150+CD48+), and fraction C (LSKCD150−CD48+) (Figure 2B). However, the subdivision of fraction A cells into MPP1 (LSKCD150+CD48−CD34+CD135−) and HSCs (LSKCD150+CD48−CD34−CD135−) uncovered a significant increase of HSCs, the most quiescent fraction of LSKs, accompanied by a decrease in the percentage of MPP1 cells in Cdk6−/− mice (Figure 2C). The enhanced representation of the most quiescent stem cells in fraction A (LSKCD150+CD48−) was accompanied by a higher expression of Tie2 (supplemental Figure 2C). Under homeostatic conditions, the majority of cells in fraction A are in the G0 phase,12 and we failed to detect any significant differences in the cell-cycle profile of Cdk6+/+ and Cdk6−/− fraction A (Figure 2D), fraction B, or fraction C cells (supplemental Figure 2D). Furthermore, we observed no significant differences in numbers of apoptotic cells in LSK and fraction A cells between genotypes (supplemental Figure 2E).
Increase in the most quiescent HSC population in Cdk6−/− mice. (A) LSK BM cells are subdivided into 3 populations based on CD150 and CD48 expression, LSKCD150+CD48− (A), LSKCD150+CD48+ (B), and LSKCD150−CD48+ (C). The LSKCD150+CD48− (A) population can be further subdivided into CD135−CD34− (HSC) and CD135−CD34+ (MPP1) subsets. Sets of representative FACS blots of Cdk6+/+ and Cdk6−/− BM cells are shown. (B) Total cell numbers of LSKCD150+CD48− (A), LSKCD150+CD48+ (B), CD150−CD48+ (C) in Cdk6+/+ and Cdk6−/− animals are shown (n = 3 per genotype). (C) Analysis of individual subpopulations of LSKCD150+CD48− (A) cells is depicted. Population A is further subdivided into HSC and MPP1 (n = 3 per genotype; *P < .05). (D) Cell-cycle distributions of fraction A cells were analyzed with DAPI and Ki-67 staining. One representative FACS blot per genotype is depicted (left). Summary of data (right; n = 6 per genotype).
Increase in the most quiescent HSC population in Cdk6−/− mice. (A) LSK BM cells are subdivided into 3 populations based on CD150 and CD48 expression, LSKCD150+CD48− (A), LSKCD150+CD48+ (B), and LSKCD150−CD48+ (C). The LSKCD150+CD48− (A) population can be further subdivided into CD135−CD34− (HSC) and CD135−CD34+ (MPP1) subsets. Sets of representative FACS blots of Cdk6+/+ and Cdk6−/− BM cells are shown. (B) Total cell numbers of LSKCD150+CD48− (A), LSKCD150+CD48+ (B), CD150−CD48+ (C) in Cdk6+/+ and Cdk6−/− animals are shown (n = 3 per genotype). (C) Analysis of individual subpopulations of LSKCD150+CD48− (A) cells is depicted. Population A is further subdivided into HSC and MPP1 (n = 3 per genotype; *P < .05). (D) Cell-cycle distributions of fraction A cells were analyzed with DAPI and Ki-67 staining. One representative FACS blot per genotype is depicted (left). Summary of data (right; n = 6 per genotype).
Enhanced lethality of Cdk6−/− mice upon repeated 5-FU treatment
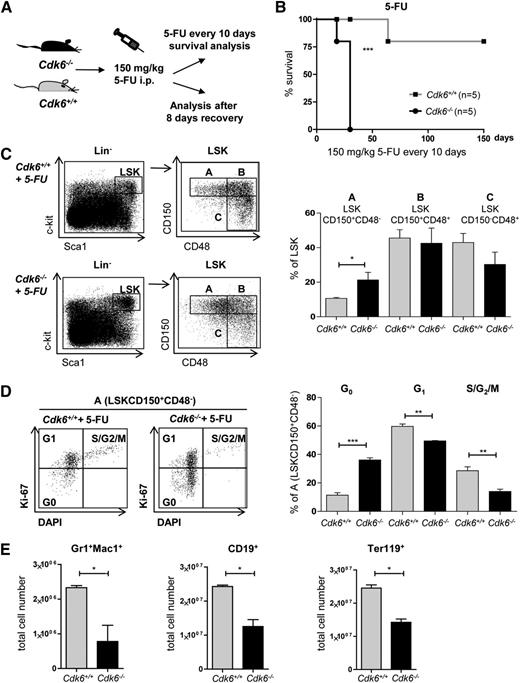
The chemotherapeutic drug 5-FU kills dividing cells but spares dormant HSCs, which are subsequently forced to proliferate to repopulate the BM.29 Cdk6−/− mice tolerated only 2 cycles of 5-FU application. In contrast, only 1 of 5 Cdk6+/+ mice succumbed to repeated 5-FU treatment within 65 days, with the remainder surviving until the experiment was terminated after 150 days (Figure 3A-B). After a single dose of 5-FU, the percentage of cells in fraction A (which includes the most quiescent stem cells) was significantly enhanced in Cdk6−/− animals within 8 days (Figure 3C). More of the cells in fraction A from the Cdk6−/− mice remained in the G0 phase (Figure 3D), indicative of their reduced ability to exit quiescence. No significant alterations in the proportion of cells in the G0 phase were detectable in cells in fractions B and C (supplemental Figure 3A). Eight days after 5-FU injection, the numbers of Gr1+Mac1+, CD19+, and Ter119+ cells were markedly lower in Cdk6−/− mice than in the control cohort (Figure 3E), but no differences in the numbers of apoptotic LSKs or fraction A cells were detectable (supplemental Figure 3B).
Impaired recovery of Cdk6−/− mice after 5-FU treatment. (A) Cdk6+/+ and Cdk6−/− mice were repeatedly treated with 150 mg/kg 5-FU every 10 days (Figure 3B) or analyzed 8 days after a single dose of 5-FU (Figure 3C-E). (B) Four of 5 Cdk6+/+ animals survived for 150 days, whereas all Cdk6−/− animals died within 25 days (n = 5 per genotype; ***P < .001). (C) Eight days after 5-FU treatment, BM of Cdk6+/+ and Cdk6−/− mice was analyzed via FACS for the presence of LSKs and LSKCD150+CD48− (A) cells. Gating strategy is shown in 1 representative FACS blot per genotype (left). Summary of statistical analyses of individual LSK subpopulations (right; n = 4 per genotype, *P < .05). (D) Cell-cycle distributions of fraction A cells were determined by a combined DAPI and Ki-67 staining. One representative FACS blot per genotype is shown (left). Summary of statistical analyses (right; n = 4 per genotype, **P < .01, ***P < .001). (E) Cdk6+/+ and Cdk6−/− mice were analyzed for lineage recovery (Gr1+Mac1+, CD19+, and Ter119+ cells) 8 days after 5-FU treatment (n = 4 per genotype; *P < .05). i.p., intraperitoneally.
Impaired recovery of Cdk6−/− mice after 5-FU treatment. (A) Cdk6+/+ and Cdk6−/− mice were repeatedly treated with 150 mg/kg 5-FU every 10 days (Figure 3B) or analyzed 8 days after a single dose of 5-FU (Figure 3C-E). (B) Four of 5 Cdk6+/+ animals survived for 150 days, whereas all Cdk6−/− animals died within 25 days (n = 5 per genotype; ***P < .001). (C) Eight days after 5-FU treatment, BM of Cdk6+/+ and Cdk6−/− mice was analyzed via FACS for the presence of LSKs and LSKCD150+CD48− (A) cells. Gating strategy is shown in 1 representative FACS blot per genotype (left). Summary of statistical analyses of individual LSK subpopulations (right; n = 4 per genotype, *P < .05). (D) Cell-cycle distributions of fraction A cells were determined by a combined DAPI and Ki-67 staining. One representative FACS blot per genotype is shown (left). Summary of statistical analyses (right; n = 4 per genotype, **P < .01, ***P < .001). (E) Cdk6+/+ and Cdk6−/− mice were analyzed for lineage recovery (Gr1+Mac1+, CD19+, and Ter119+ cells) 8 days after 5-FU treatment (n = 4 per genotype; *P < .05). i.p., intraperitoneally.
Reduced activation of dormant Cdk6−/− HSCs after poly(I:C) treatment
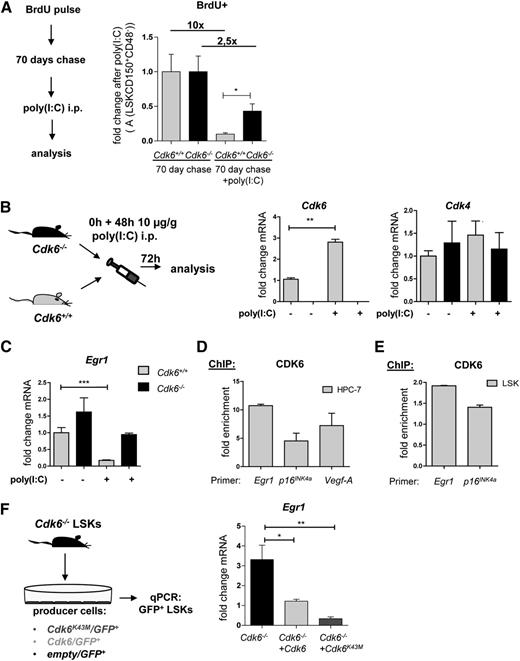
These findings indicate a possible malfunction of the most quiescent stem cells in fraction A of Cdk6−/− mice. To test this notion, we performed in vivo label-retaining experiments using 5-bromo-2′-deoxyuridine (BrdU). After a 70-day chase period, the most quiescent stem cell fraction remained BrdU-positive.12,30 When we provoked HSC proliferation by injecting poly(I:C),31 significantly more cells remained BrdU-positive in Cdk6−/− mice (Figure 4A), indicative of their restricted ability to leave quiescence. Experiments without prior BrdU labeling confirmed that significantly more cells of fraction A in Cdk6−/− mice remain in the G0 phase (supplemental Figure 4A-B). No alterations in the cell-cycle distribution of fraction B or C cells were detected (supplemental Figure 4C).
Impaired activation of dormant Cdk6−/− HSCs after poly(I:C) treatment. (A) Cdk6+/+ and Cdk6−/− mice received a pulse of BrdU i.p. (1 mg/mouse) that was followed by a 10-day period of BrdU administration via drinking water (1 mg/mL) (n = 8 per genotype). After a 70-day chase period, the mice cohort was split and BrdU+ fraction A cells were analyzed with (n = 4 per genotype) or without (n = 4 per genotype) a 24-hour preceding injection of poly(I:C) (10 µg/g body weight). Treatment with poly(I:C) resulted in a 10-fold reduction of BrdU+ fraction A cells in Cdk6+/+, but only in a 2.5-fold reduction in Cdk6−/− mice. (B) Cdk6 and Cdk4 mRNA expression levels in fraction A cells of Cdk6+/+ and Cdk6−/− mice that had received poly(I:C) or PBS are shown (n = 3 for each genotype; qPCR analyses were performed in triplicate; **P < .01). (C) Egr1 mRNA expression levels in fraction A cells of Cdk6+/+ and Cdk6−/− mice that had received poly(I:C) or PBS are shown (n = 6 for each genotype; qPCR analyses were performed in triplicate; ***P < .001). (D-E) ChIP assays were performed in an HPC-7 hematopoietic progenitor cell line (D) and in primary Cdk6+/+ LSKs (E). Protein–DNA complexes were immunoprecipitated using antibodies directed against CDK6 and analyzed by qPCR for their presence on the Egr1 promoter region. Vegf-A and/or p16INK4a promoter regions were used as positive controls. Bar graphs depict fold enrichment over a negative region downstream of CD19. (F) Cdk6−/− LSKs were sorted and coincubated with Cdk6/GFP+, Cdk6K43M/GFP+, or empty/GFP+ GP+ producer cells (n = 3 per genotype). After 48 hours, GFP+ cells were high-purity sorted by FACS and analyzed by qPCR. Bar graphs depict Egr1 mRNA expression levels (technical triplicates; *P < .05, **P < .01).
Impaired activation of dormant Cdk6−/− HSCs after poly(I:C) treatment. (A) Cdk6+/+ and Cdk6−/− mice received a pulse of BrdU i.p. (1 mg/mouse) that was followed by a 10-day period of BrdU administration via drinking water (1 mg/mL) (n = 8 per genotype). After a 70-day chase period, the mice cohort was split and BrdU+ fraction A cells were analyzed with (n = 4 per genotype) or without (n = 4 per genotype) a 24-hour preceding injection of poly(I:C) (10 µg/g body weight). Treatment with poly(I:C) resulted in a 10-fold reduction of BrdU+ fraction A cells in Cdk6+/+, but only in a 2.5-fold reduction in Cdk6−/− mice. (B) Cdk6 and Cdk4 mRNA expression levels in fraction A cells of Cdk6+/+ and Cdk6−/− mice that had received poly(I:C) or PBS are shown (n = 3 for each genotype; qPCR analyses were performed in triplicate; **P < .01). (C) Egr1 mRNA expression levels in fraction A cells of Cdk6+/+ and Cdk6−/− mice that had received poly(I:C) or PBS are shown (n = 6 for each genotype; qPCR analyses were performed in triplicate; ***P < .001). (D-E) ChIP assays were performed in an HPC-7 hematopoietic progenitor cell line (D) and in primary Cdk6+/+ LSKs (E). Protein–DNA complexes were immunoprecipitated using antibodies directed against CDK6 and analyzed by qPCR for their presence on the Egr1 promoter region. Vegf-A and/or p16INK4a promoter regions were used as positive controls. Bar graphs depict fold enrichment over a negative region downstream of CD19. (F) Cdk6−/− LSKs were sorted and coincubated with Cdk6/GFP+, Cdk6K43M/GFP+, or empty/GFP+ GP+ producer cells (n = 3 per genotype). After 48 hours, GFP+ cells were high-purity sorted by FACS and analyzed by qPCR. Bar graphs depict Egr1 mRNA expression levels (technical triplicates; *P < .05, **P < .01).
Exit from quiescence is controlled by a network of transcriptional regulators including Nurr1, Egr1, Runx1, and p21CIP1. Poly(I:C) treatment induced Cdk6 expression by about 3-fold in the cells of fraction A, whereas the level of Cdk4 messenger RNA (mRNA) remained unchanged (Figure 4B). There were no changes in the levels of other transcription factors implicated in the regulation of stem cell quiescence (p16INK4A, p53, p27KIP1, p21CIP1, Smad3, Smad4, Smad7, Foxo1a, JunB, and Runx1; supplemental Figure 5A). Consistent with previous findings,5 we detected altered Notch1 regulation (supplemental Figure 5A). Furthermore, we observed increased Nurr1 expression in fraction A cells of nonstimulated Cdk6−/− mice, but no differences between Cdk6+/+ and Cdk6−/− mice in the levels of Nurr1 RNA in stimulated cells of fraction A. The changes in expression of Notch132 and Nurr1 are insufficient to account for the phenotype of Cdk6−/− HSCs. We found a pronounced downregulation of Egr1 upon poly(I:C) treatment of fraction A cells of Cdk6+/+ mice that was not observed in fraction A cells of Cdk6−/− mice (Figure 4C). Downregulation of Egr1 is necessary for HSCs to exit quiescence.13
CDK6 directly regulates Egr1 expression in hematopoietic cells
Poly(I:C) stimulation induces Cdk6 and suppresses Egr1, suggesting that CDK6 may directly regulate Egr1 expression. We have identified a transcriptional role for CDK6 in lymphoid malignancies7 and so we used BCR-ABLp185+–transformed pro-B cells for initial investigations of the Egr1 promoter. Egr1 is expressed at high levels in transformed differentiated cells and functions as a proto-oncogene in certain tumor types.33-35 Consistently, we find high levels of Cdk6 mRNA paralleled by a statistically significant upregulation of Egr1 in transformed BCR-ABLp185+ cells (supplemental Figure 6A). Chromatin immunoprecipitation (ChIP) analysis revealed the presence of CDK6 at the Egr1 promoter at levels comparable to its binding to the p16INK4A and Vegf-A promoters, which are known transcriptional targets of CDK6 in this cell type (supplemental Figure 6B).7 ChIP assays with the hematopoietic progenitor cell line HPC-7 and with sorted LSKs (with Vegf-A and/or p16INK4A promoters as positive controls) showed that CDK6 also binds to the Egr1 promoter in these cell types (Figure 4D-E).
CDK6 has recently been shown to influence transcription independently of its kinase function. To examine whether CDK6 activates Egr1 in a kinase-dependent or kinase-independent manner, we used the kinase-dead mutant Cdk6K43M. ChIP assays confirmed that CDK6K43M binds to the Egr1 promoter in BCR-ABLp185+ pro-B cells (supplemental Figure 6C). Expression of Cdk6K43M significantly regulated Egr1 levels in BCR-ABLp185+ pro-B cells (supplemental Figure 6D) and in Cdk6−/− LSKs transduced with Cdk6, Cdk6K43M, or empty vector control (Figure 4F; supplemental Figure 6E).
CDK6 is part of a transcription factor network that regulates stem cell quiescence
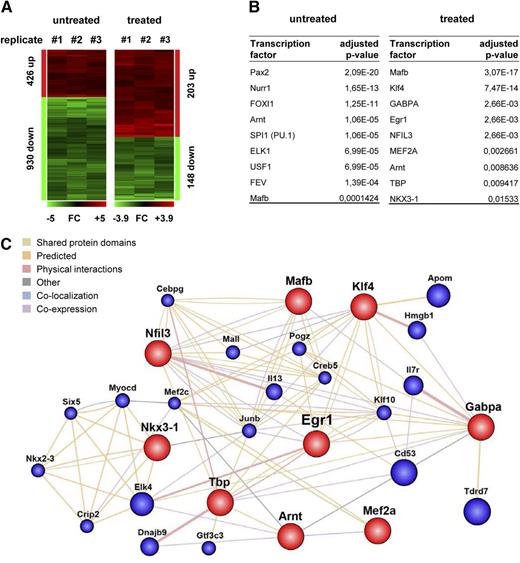
To investigate the effects of poly(I:C) stimulation of Cdk6+/+ and Cdk6−/− cells in fraction A, we performed transcriptional profiling as described previously.23 Untreated fraction A cells of Cdk6−/− mice had greater levels of 426 protein-coding transcripts and lower levels of 930 protein-coding transcripts compared with cells from Cdk6+/+ mice. Upon poly(I:C) stimulation, differences between the genotypes were found in 203 (upregulated) and 148 (downregulated) protein-coding transcripts (Figure 5A, supplemental Tables 1 and 2). Gene ontology analysis suggested that the deregulated genes are involved in a variety of biological processes, as summarized in Table 1.
Transcriptional profiling of poly(I:C) treated and untreated Cdk6−/− HSCs. (A) A summary of transcripts up- (red) or down- (green) regulated (fold-change >2) in fraction A cells that were FACS-purified from either untreated or poly(I:C)-treated Cdk6−/− and Cdk6+/+ mice (n = 3 per genotype). (B) Transcripts deregulated in either untreated or poly(I:C)-treated Cdk6−/− fraction A cells displayed promoter sequences significantly enriched in recognition sites for the indicated transcription factors compared with the respective Cdk6+/+ controls. (C) GeneMANIA-computed association network of the transcription factors identified via their deregulated target gene programs in poly(I:C)-treated Cdk6−/− fraction A cells vs Cdk6+/+ controls.
Transcriptional profiling of poly(I:C) treated and untreated Cdk6−/− HSCs. (A) A summary of transcripts up- (red) or down- (green) regulated (fold-change >2) in fraction A cells that were FACS-purified from either untreated or poly(I:C)-treated Cdk6−/− and Cdk6+/+ mice (n = 3 per genotype). (B) Transcripts deregulated in either untreated or poly(I:C)-treated Cdk6−/− fraction A cells displayed promoter sequences significantly enriched in recognition sites for the indicated transcription factors compared with the respective Cdk6+/+ controls. (C) GeneMANIA-computed association network of the transcription factors identified via their deregulated target gene programs in poly(I:C)-treated Cdk6−/− fraction A cells vs Cdk6+/+ controls.
Gene ontology analysis of transcripts deregulated in untreated and poly(I:C) treated Cdk6−/− fraction A cells
| GO ID . | Description . | Representative genes upregulated . | Representative genes downregulated . |
|---|---|---|---|
| Untreated | |||
| GO:0070887 | Cellular response to chemical stimulus | Arnt, Cntf, Egr1, Ern1, Esr1, Etv5, Kif16b, Nr4a2, Rabgef1, Skil, Smad7 | Aicda, Bcar1, Bmp2, Ccl6, Cxcr2, Lyst, Ngfr, Pax2, Tbx1, Timp2, Trib1 |
| GO:0098552 | Side of membrane | Amot, Cxcl9, Dlk1, Folr2,Hyal2, Il12rb1, Klre1 | Cd74, Cd80, Ceacam2, Efna5, Fas, Fcer1a, Itga1, Rasa2 |
| GO:0009605 | Response to external stimulus | Bmp7, Chn1, Gbp6, Nr4a2, Rab38 | Ablim1, Cxcr1, L1cam, Ngfr, Penk, Slit3 |
| GO:0031012 | Extracellular matrix | Chad, Col16a1, Col18a1, Dspp, Prss12 | Ache, Gpc3, Leprel1, Lox, Lpl, Otog, Serac1, Vcan |
| GO:0048583 | Regulation of response to stimulus | Amot, Egr1, Klre1, Rabgef1, Sybu | Acp5, Apod, Disc1, Fcgr3, Notch1, Slit3 |
| GO:0009986 | Cell surface | Cxcl9, Dlk1, Folr2, H2-K1, Klre1 | Ache, Ceacam2, Clec7a, Efna5, Gfra1, Ramp1 |
| GO:0043235 | Receptor complex | Il12rb1, Rnmt | Cacng3, Card11, Egfr, Gpr160, Itga1 |
| GO:0031224 | Intrinsic component of membrane | Apool, B4galt7, Gpr125, Plscr4, Zdhhc20 | Ache, Clcc1, Grina, Layn, Scn11a, Tmem160 |
| GO:0032502 | Developmental process | Catsper4, Dpf1, Nanog, Rbm19, Tshz1 | Antxr1, Cdh4, Dkkl1, E2f7, Elk1, Tg |
| GO:0005615 | Extracellular space | C1rl, Cntf, Eef1a1, Fam20c, Retn | Acta2, Cfb, Ctsk, Inhba, Lepr |
| GO:0040011 | Locomotion | Bmp7, Hyal2, Nr4a1, P2ry1 | Ablim1, Cemip, Dock4, L1cam, Snai2 |
| Poly:IC treated | |||
| GO:0070887 | Cellular response to chemical stimulus | Ahr, Ctr9, Dgat2, Egr1, Egr2, Egr3, Klf4, Nfil3 | Ackr4, Fam132a, Gcg, Phip, Pklr |
| GO:0001071 | Nucleic acid binding transcription factor activity | Btg2, Fosl2, Hes1, Id3, Mafb | E2f7, Foxl2, Hmga1, Nkx2-2, Zfp628 |
| GO:0016265 | Death | Adamtsl4, Hcar2, Lgmn, Perp, Xaf1 | Atm, Gcg, Optn, Phip, Wrn |
| GO:0010033 | Response to organic substance | Dgat2, Egr1, Egr2, Egr3, Fosl2, Mgst1 | Fgf8, Gpd1, Hpx, Iigp1, Pklr |
| GO:0003677 | DNA binding | Csrnp1, Hes1, Hist1h1c, Nfil3, Tbx3 | E2f7, Jrk, Sox6, Zfp518a, Zfp628 |
| GO ID . | Description . | Representative genes upregulated . | Representative genes downregulated . |
|---|---|---|---|
| Untreated | |||
| GO:0070887 | Cellular response to chemical stimulus | Arnt, Cntf, Egr1, Ern1, Esr1, Etv5, Kif16b, Nr4a2, Rabgef1, Skil, Smad7 | Aicda, Bcar1, Bmp2, Ccl6, Cxcr2, Lyst, Ngfr, Pax2, Tbx1, Timp2, Trib1 |
| GO:0098552 | Side of membrane | Amot, Cxcl9, Dlk1, Folr2,Hyal2, Il12rb1, Klre1 | Cd74, Cd80, Ceacam2, Efna5, Fas, Fcer1a, Itga1, Rasa2 |
| GO:0009605 | Response to external stimulus | Bmp7, Chn1, Gbp6, Nr4a2, Rab38 | Ablim1, Cxcr1, L1cam, Ngfr, Penk, Slit3 |
| GO:0031012 | Extracellular matrix | Chad, Col16a1, Col18a1, Dspp, Prss12 | Ache, Gpc3, Leprel1, Lox, Lpl, Otog, Serac1, Vcan |
| GO:0048583 | Regulation of response to stimulus | Amot, Egr1, Klre1, Rabgef1, Sybu | Acp5, Apod, Disc1, Fcgr3, Notch1, Slit3 |
| GO:0009986 | Cell surface | Cxcl9, Dlk1, Folr2, H2-K1, Klre1 | Ache, Ceacam2, Clec7a, Efna5, Gfra1, Ramp1 |
| GO:0043235 | Receptor complex | Il12rb1, Rnmt | Cacng3, Card11, Egfr, Gpr160, Itga1 |
| GO:0031224 | Intrinsic component of membrane | Apool, B4galt7, Gpr125, Plscr4, Zdhhc20 | Ache, Clcc1, Grina, Layn, Scn11a, Tmem160 |
| GO:0032502 | Developmental process | Catsper4, Dpf1, Nanog, Rbm19, Tshz1 | Antxr1, Cdh4, Dkkl1, E2f7, Elk1, Tg |
| GO:0005615 | Extracellular space | C1rl, Cntf, Eef1a1, Fam20c, Retn | Acta2, Cfb, Ctsk, Inhba, Lepr |
| GO:0040011 | Locomotion | Bmp7, Hyal2, Nr4a1, P2ry1 | Ablim1, Cemip, Dock4, L1cam, Snai2 |
| Poly:IC treated | |||
| GO:0070887 | Cellular response to chemical stimulus | Ahr, Ctr9, Dgat2, Egr1, Egr2, Egr3, Klf4, Nfil3 | Ackr4, Fam132a, Gcg, Phip, Pklr |
| GO:0001071 | Nucleic acid binding transcription factor activity | Btg2, Fosl2, Hes1, Id3, Mafb | E2f7, Foxl2, Hmga1, Nkx2-2, Zfp628 |
| GO:0016265 | Death | Adamtsl4, Hcar2, Lgmn, Perp, Xaf1 | Atm, Gcg, Optn, Phip, Wrn |
| GO:0010033 | Response to organic substance | Dgat2, Egr1, Egr2, Egr3, Fosl2, Mgst1 | Fgf8, Gpd1, Hpx, Iigp1, Pklr |
| GO:0003677 | DNA binding | Csrnp1, Hes1, Hist1h1c, Nfil3, Tbx3 | E2f7, Jrk, Sox6, Zfp518a, Zfp628 |
Most significantly deregulated functional pathways and their Gene Ontology (GO) codes are shown first.
The majority of the known regulators of stem cell quiescence were not significantly affected, although there were significant changes to the levels of Nurr1, Atm, Hoxc4, and (especially) Egr1 (Table 2). The outcome of a JASPAR-TFB screen for targets of specific transcription factors is summarized in Figure 5B. We found levels of Nurr1 target genes (supplemental Table 3) to be significantly affected by the loss of CDK6 in unstimulated cells. In contrast, the key transcription factors with different activities in stimulated Cdk6−/− and Cdk6+/+ cells included Egr1 and Klf4. Egr1 has been reported to regulate Klf4,36 so we used GeneMANIA to investigate further interconnections between the individual regulators identified by JASPAR-TFB (Figure 5C). Array hit validation by quantitative polymerase chain reaction (qPCR) of individual genes found to be deregulated in untreated or poly(I:C)-treated cells of Cdk6−/− fraction A (Klf4, PU.1, Arnt) were consistent with our data set (supplemental Figure 7A). In summary, the findings indicate that the CDK6-Egr1 axis represents an important part of a transcription factor network that controls exit from quiescence of HSCs.
Regulators of HSC quiescence
| Gene symbol . | Description . | Fold-change untreated . | P value untreated . | Regulation untreated . | Fold-change poly:IC . | P value poly:IC . | Regulation poly:IC . |
|---|---|---|---|---|---|---|---|
| Egr1 | Early growth response 1 | 2.90 | 2.11E-03 | Up | 2.62 | 1.25E-02 | Up |
| Atm | Ataxia telangiectasia mutated homolog (human) | 1.50 | 6.02E-01 | — | 2.08 | 4.62E-02 | Down |
| Nurr1 | Nuclear receptor subfamily 4, group A, member 2, transcript variant 1 | 4.03 | 1.01E-03 | Up | 2.68 | 7.45E-02 | — |
| Cdkn2c | CDK inhibitor 2C (p18, inhibits CDK4) | 2.21 | 2.36E-01 | — | 2.16 | 1.71E-01 | — |
| Gfi1 | Growth factor independent 1 | 1.12 | 6.56E-01 | — | 1.96 | 2.77E-02 | — |
| Stat1 | Signal transducer and activator of transcription 1, transcript variant 2 | 1.07 | 6.48E-01 | — | 1.75 | 2.24E-02 | — |
| Hoxb9 | Homeobox B9 | 1.27 | 4.77E-01 | — | 1.70 | 4,52E-02 | — |
| Junb | Jun B proto-oncogene | 1.21 | 2.92E-01 | — | 1.65 | 8,99E-02 | — |
| Pml | Promyelocytic leukemia, transcript variant 2 | 1.22 | 8.64E-02 | — | 1.64 | 4.62E-02 | — |
| Foxo4 | Forkhead box O4 | 1.75 | 1.89E-03 | — | 1.54 | 2.93E-01 | — |
| Hoxa4 | Homeobox A4 | 1.03 | 8.27E-01 | — | 1.47 | 3.07E-01 | — |
| Bmi1 | Bmi1 polycomb ring finger oncogene | 1.21 | 1.38E-01 | — | 1.38 | 2.76E-01 | — |
| Trp53 | Transformation related protein 53 | 1.52 | 1.81E-02 | — | 1.35 | 1.26E-01 | — |
| Txnip | Thioredoxin interacting protein | 1.75 | 3.40E-02 | — | 1.27 | 2.42E-01 | — |
| Shh | Sonic hedgehog | 1.25 | 2.30E-02 | — | 1.26 | 2.44E-01 | — |
| Tal1 | T-cell acute lymphocytic leukemia 1, transcript variant 2 | 1.07 | 6.91E-01 | — | 1.25 | 1.93E-01 | — |
| Satb1 | Special AT-rich sequence binding protein 1, transcript variant 2 | 1.65 | 1.71E-01 | — | 1.22 | 4.03E-01 | — |
| Hoxc4 | Homeobox C4 | 6.78 | 8.87E-03 | Up | 1.19 | 7.98E-01 | — |
| Itch | Itchy, E3 ubiquitin protein ligase, transcript variant 2 | 1.21 | 2.00E-01 | — | 1.14 | 7.27E-01 | — |
| Pbx1 | Pre B-cell leukemia homeobox 1 (Pbx1), transcript variant a | 1.27 | 7.09E-02 | — | 1.12 | 3.57E-01 | — |
| Gene symbol . | Description . | Fold-change untreated . | P value untreated . | Regulation untreated . | Fold-change poly:IC . | P value poly:IC . | Regulation poly:IC . |
|---|---|---|---|---|---|---|---|
| Egr1 | Early growth response 1 | 2.90 | 2.11E-03 | Up | 2.62 | 1.25E-02 | Up |
| Atm | Ataxia telangiectasia mutated homolog (human) | 1.50 | 6.02E-01 | — | 2.08 | 4.62E-02 | Down |
| Nurr1 | Nuclear receptor subfamily 4, group A, member 2, transcript variant 1 | 4.03 | 1.01E-03 | Up | 2.68 | 7.45E-02 | — |
| Cdkn2c | CDK inhibitor 2C (p18, inhibits CDK4) | 2.21 | 2.36E-01 | — | 2.16 | 1.71E-01 | — |
| Gfi1 | Growth factor independent 1 | 1.12 | 6.56E-01 | — | 1.96 | 2.77E-02 | — |
| Stat1 | Signal transducer and activator of transcription 1, transcript variant 2 | 1.07 | 6.48E-01 | — | 1.75 | 2.24E-02 | — |
| Hoxb9 | Homeobox B9 | 1.27 | 4.77E-01 | — | 1.70 | 4,52E-02 | — |
| Junb | Jun B proto-oncogene | 1.21 | 2.92E-01 | — | 1.65 | 8,99E-02 | — |
| Pml | Promyelocytic leukemia, transcript variant 2 | 1.22 | 8.64E-02 | — | 1.64 | 4.62E-02 | — |
| Foxo4 | Forkhead box O4 | 1.75 | 1.89E-03 | — | 1.54 | 2.93E-01 | — |
| Hoxa4 | Homeobox A4 | 1.03 | 8.27E-01 | — | 1.47 | 3.07E-01 | — |
| Bmi1 | Bmi1 polycomb ring finger oncogene | 1.21 | 1.38E-01 | — | 1.38 | 2.76E-01 | — |
| Trp53 | Transformation related protein 53 | 1.52 | 1.81E-02 | — | 1.35 | 1.26E-01 | — |
| Txnip | Thioredoxin interacting protein | 1.75 | 3.40E-02 | — | 1.27 | 2.42E-01 | — |
| Shh | Sonic hedgehog | 1.25 | 2.30E-02 | — | 1.26 | 2.44E-01 | — |
| Tal1 | T-cell acute lymphocytic leukemia 1, transcript variant 2 | 1.07 | 6.91E-01 | — | 1.25 | 1.93E-01 | — |
| Satb1 | Special AT-rich sequence binding protein 1, transcript variant 2 | 1.65 | 1.71E-01 | — | 1.22 | 4.03E-01 | — |
| Hoxc4 | Homeobox C4 | 6.78 | 8.87E-03 | Up | 1.19 | 7.98E-01 | — |
| Itch | Itchy, E3 ubiquitin protein ligase, transcript variant 2 | 1.21 | 2.00E-01 | — | 1.14 | 7.27E-01 | — |
| Pbx1 | Pre B-cell leukemia homeobox 1 (Pbx1), transcript variant a | 1.27 | 7.09E-02 | — | 1.12 | 3.57E-01 | — |
Summary of HSC quiescence regulating genes that are differently regulated in Cdk6−/− fraction A cells (listed in order of “fold-change”) in steady state (“untreated”) or upon in vivo poly(I:C) stimulation. In the absence of CDK6, Egr1 levels are significantly higher compared with control. This difference is even more prominent in the poly(I:C)-treated fraction, in which Egr1 represents the most deregulated factor. Statistical significance (fold-change >2; P < .05).
The key role of CDK6 in regulating stem cell quiescence extends to LSCs
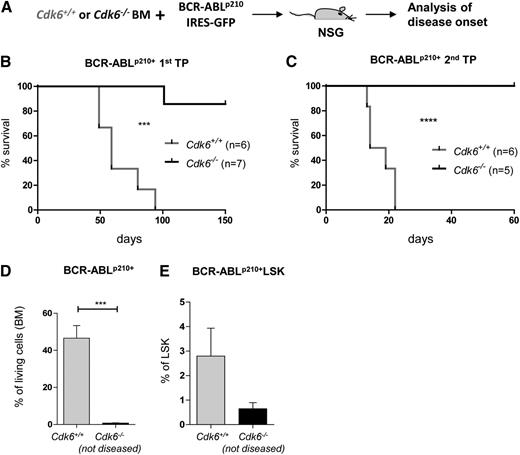
HSCs represent the LSCs’ compartment in BCR-ABLp210+–induced leukemia.37 To investigate whether CDK6 regulates Egr1 in LSCs, we infected Cdk6+/+ and Cdk6−/− BM cells with a retrovirus encoding BCR-ABLp210-IRES GFP and injected them into nonirradiated NSG mice (Figure 6A). This procedure triggers a CML-like disease that depends on the constant replenishment of peripheral leukemic cells by BCR-ABLp210+ LSCs. In line with published results,37 all mice that received BCR-ABLp210+Cdk6+/+ BM succumbed to disease within 3 months, whereas only 1 of the 7 animals that received BCR-ABLp210+Cdk6−/− cells became sick within this period (Figure 6B). Differences were even more explicit in a second round of transplantation, which again forced the BCR-ABLp210+ LSCs to repopulate. Although comparable amounts of BCR-ABLp210+ BM cells were used for transplantation, only mice that received Cdk6+/+ leukemic cells succumbed to leukemia (Figure 6C). Mice that received BCR-ABLp210+Cdk6−/− cells remained disease-free for up to 60 days. Analysis of the mice confirmed that Cdk6−/− LSCs had homed efficiently to the BM. Despite their presence, no signs of leukemia could be detected in the recipient animals (Figure 6D-E). BCR-ABLp210+ LSCs derived from Cdk6+/+ and Cdk6−/− mice express identical surface markers, which are reminiscent of those on fraction C cells (supplemental Figure 8A). When we analyzed the cell cycle of the remaining Cdk6+/+ and Cdk6−/− BCR-ABLp210+ LSKs, we again found more Cdk6−/− BCR-ABLp210+ LSKs in the G0 phase (supplemental Figure 8B).
CDK6 is required for leukemia formation in vivo. (A) Experimental setup: Cdk6+/+ and Cdk6−/− BM cells were cocultivated on BCR-ABLp210 producer cells and 2 × 106 cells were injected i.v. in nonirradiated NSG mice. (B) Kaplan-Meier plot depicting disease onset of NSGs injected with Cdk6+/+ or Cdk6−/− BCR-ABLp210+ leukemic cells (n = 6 and n = 7, respectively). Only 1 of 7 mice injected with Cdk6−/− BCR-ABLp210+ cells became diseased. All mice injected with Cdk6+/+ BCR-ABLp210+ cells became diseased within 3 months (***P < .001). (C) 2 × 106 BM cells of diseased animals were transplanted in a second transplantation round and disease onset was monitored. None of the mice injected with Cdk6−/− BCR-ABLp210+ cells became diseased, but all mice injected with Cdk6+/+ BCR-ABLp210+ cells became diseased rapidly within 3 weeks (n = 5 and n = 6, respectively; ****P < .0001). (D) The experiment (Figure 6C) was terminated after 60 days and Cdk6−/− nondiseased animals were euthanized. Contribution of Cdk6−/− BCR-ABLp210+–transformed cells (BM) was compared with diseased control animals at the time of terminal disease (n = 3 per genotype, ***P < .001). (E) Frequencies of BM BCR-ABLp210+ LSKs of Cdk6−/− animals compared with Cdk6+/+ (diseased) animals. Cdk6−/− BCR-ABLp210+ LSKs were detectable in the BM (n = 3 per genotype).
CDK6 is required for leukemia formation in vivo. (A) Experimental setup: Cdk6+/+ and Cdk6−/− BM cells were cocultivated on BCR-ABLp210 producer cells and 2 × 106 cells were injected i.v. in nonirradiated NSG mice. (B) Kaplan-Meier plot depicting disease onset of NSGs injected with Cdk6+/+ or Cdk6−/− BCR-ABLp210+ leukemic cells (n = 6 and n = 7, respectively). Only 1 of 7 mice injected with Cdk6−/− BCR-ABLp210+ cells became diseased. All mice injected with Cdk6+/+ BCR-ABLp210+ cells became diseased within 3 months (***P < .001). (C) 2 × 106 BM cells of diseased animals were transplanted in a second transplantation round and disease onset was monitored. None of the mice injected with Cdk6−/− BCR-ABLp210+ cells became diseased, but all mice injected with Cdk6+/+ BCR-ABLp210+ cells became diseased rapidly within 3 weeks (n = 5 and n = 6, respectively; ****P < .0001). (D) The experiment (Figure 6C) was terminated after 60 days and Cdk6−/− nondiseased animals were euthanized. Contribution of Cdk6−/− BCR-ABLp210+–transformed cells (BM) was compared with diseased control animals at the time of terminal disease (n = 3 per genotype, ***P < .001). (E) Frequencies of BM BCR-ABLp210+ LSKs of Cdk6−/− animals compared with Cdk6+/+ (diseased) animals. Cdk6−/− BCR-ABLp210+ LSKs were detectable in the BM (n = 3 per genotype).
Egr1 knock-down rescues colony formation in Cdk6−/− BCR-ABLp210+ LSCs
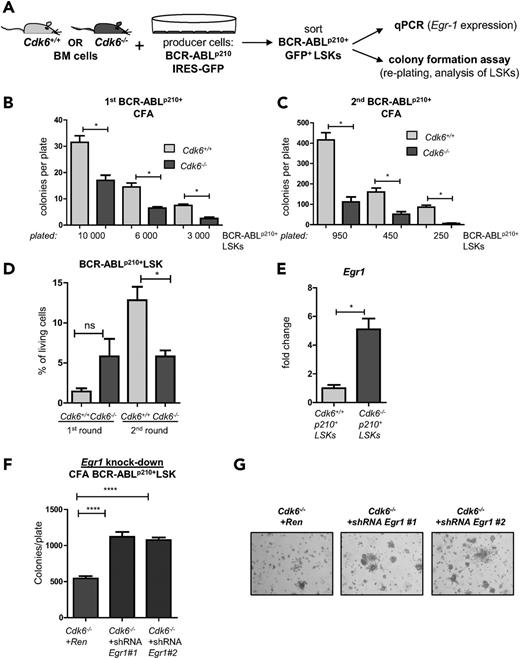
In vitro colony formation assay confirmed the in vivo observation. After 48 hours of coculture of BM cells on retroviral producers, BCR-ABLp210+ LSKs were sorted and either seeded at 3 different cell numbers in methylcellulose to assess their (re-)plating potential or used for Egr1 qPCR (Figure 7A). Eight days after plating, we observed a profound reduction of Cdk6−/− leukemic clones, irrespective of the number of cells initially plated (Figure 7B-C). After each , all colonies were harvested to determine the number of BCR-ABLp210+ LSK cells. Whereas LSK numbers increased in BCR-ABLp210+Cdk6+/+ cells, the numbers of Cdk6−/− BCR-ABLp210+ LSKs remained stable (Figure 7D). These observations confirm the crucial role of CDK6 in BCR-ABLp210+ LSCs.
CDK6 influences (re-)plating capacities of BCR-ABLp210+ LSKs in vitro. (A) Experimental setup: Cdk6+/+ and Cdk6−/− BM cells were cocultivated on BCR-ABLp210 producer cells for 48 hours, sorted by high-purity FACS, and either subjected to colony formation assays (CFA) (B-D) or analyzed by qPCR (E). (B) Three different cell numbers of BCR-ABLp210+ LSKs were seeded and colony numbers were counted 8 days after coculture (technical duplicates; *P < .05). (C) All colonies were harvested and reseeded to a second round of replating. Colonies were counted after 8 days (technical duplicates; *P < .05). (D) After each round, colonies were harvested and analyzed by FACS for the presence of remaining BCR-ABLp210+ LSKs (*P < .05). (E) BCR-ABLp210+ LSKs were sorted by FACS and Egr1 expression was analyzed by qPCR (BM cells of 3 individual mice per genotype were pooled; qPCR was performed in technical triplicates; *P < .05). (F) Knockdown constructs Egr1 #1 and Egr1 #2 or a control vector targeting Renilla were introduced into Cdk6−/− BCR-ABLp210+ LSKs and subjected to colony formation. Colonies were again counted 8 days after seeding (****P < .0001). (G) Representative pictures of colonies on day 8 (magnification: ×4).
CDK6 influences (re-)plating capacities of BCR-ABLp210+ LSKs in vitro. (A) Experimental setup: Cdk6+/+ and Cdk6−/− BM cells were cocultivated on BCR-ABLp210 producer cells for 48 hours, sorted by high-purity FACS, and either subjected to colony formation assays (CFA) (B-D) or analyzed by qPCR (E). (B) Three different cell numbers of BCR-ABLp210+ LSKs were seeded and colony numbers were counted 8 days after coculture (technical duplicates; *P < .05). (C) All colonies were harvested and reseeded to a second round of replating. Colonies were counted after 8 days (technical duplicates; *P < .05). (D) After each round, colonies were harvested and analyzed by FACS for the presence of remaining BCR-ABLp210+ LSKs (*P < .05). (E) BCR-ABLp210+ LSKs were sorted by FACS and Egr1 expression was analyzed by qPCR (BM cells of 3 individual mice per genotype were pooled; qPCR was performed in technical triplicates; *P < .05). (F) Knockdown constructs Egr1 #1 and Egr1 #2 or a control vector targeting Renilla were introduced into Cdk6−/− BCR-ABLp210+ LSKs and subjected to colony formation. Colonies were again counted 8 days after seeding (****P < .0001). (G) Representative pictures of colonies on day 8 (magnification: ×4).
Egr1 levels were significantly higher in Cdk6−/− BCR-ABLp210+ LSKs than in Cdk6+/+ BCR-ABLp210+ LSKs (Figure 7E). To assess the functional relevance, we performed short hairpin RNA–mediated knockdowns for Egr1, validating the constructs in a hematopoietic cell line (BCR-ABLp185+ pro B cells) (supplemental Figure 9A) before introducing them into Cdk6−/− BCR-ABLp210+ LSKs. Colony formation assays revealed a highly significant increase in growth factor–independent colony numbers (Figure 7F-G) and thus confirmed the importance of the CDK6/Egr1 axis in BCR-ABLp210+–mediated leukemogenesis.
Discussion
HSC homeostasis requires the precise regulation of cell proliferation because the maintenance of long-term repopulation capacity is crucial for the ability to produce blood cells. Dormant HSCs represent a reservoir for hematopoiesis and are ready to be rapidly activated when required.16 We identified a dual role for CDK6 in HSCs—in addition to its function as a cell-cycle kinase, CDK6 downregulates Egr1 by directly binding to its promoter. This important function is also performed in LSCs, because Cdk6−/− BCR-ABLp210+-transformed LSKs are incapable of inducing disease.
We failed to detect any changes in LSK populations in young Cdk6−/− mice under steady-state conditions. This finding is consistent with the initial results of Hu et al,5 although the group subsequently38 reported slightly reduced numbers of LSKs in Cdk6−/− mice. It is conceivable that the minor differences reported in the second article arise as a result of the knockout strategy employed or from differences in housing conditions. We are not able to explain this discrepancy; our results are fully consistent with the earlier article from Hu et al.
CDK6 has a unique role under stress conditions when d-HSCs are forced to exit G0. CDK6 is not required under steady-state conditions, when CDK4 alone is sufficient to drive proliferation. Cdk6 is upregulated in HSCs under stress conditions, showing that it has an important role when a rapid and fast response of HSCs is required. Cdk4 expression remains constant, and we failed to detect any compensatory upregulation in Cdk6−/− animals.
We have recently shown that CDK6 is not only a cell-cycle kinase but also a direct regulator of transcription.7 Deleting CDK6 did not result in consistent differences in the levels of known cell-cycle regulators such as p21Cdkn1a, p27Cdkn1b, and p16INK4a or of in the transcription factors p53, JunB, Runx1, and members of the transforming growth factor-β (TGF-β) pathway. Hu et al have reported altered regulation of Notch target genes in Cdk6−/− T cells5 ; we have confirmed this finding in HSCs and show that the mRNA levels of Notch1 depend on CDK6 both under steady-state conditions as well as upon poly(I:C) treatment. Moreover, we find deregulation of Nurr1, a nuclear receptor and transcription factor that is known to be involved in HSC quiescence. HSCs overexpressing Nurr1 are capable of homing but fail to replenish the blood system.39 Significant differences in Nurr1 expression were only found in nonstimulated HSCs, where Nurr1 was the dominant transcriptional regulator that distinguishes Cdk6−/− HSCs from controls. The difference is no longer as pronounced in stimulated HSCs, where downregulation of Nurr1 occurs independently of CDK6. Effects on Nurr1 thus do not correspond to the phenotype of CDK6 deletion, confirming that other mechanisms are involved.
All of our lines of investigation are consistent with the idea that the effects of CDK6 deletion are mediated to a large extent by the regulation of Egr1. In quiescent HSCs, Egr1 is expressed at high levels and it must be downregulated to enable cells to proliferate upon stress.13 CDK6 is part of the transcriptional apparatus that suppresses Egr1. CDK6 thus performs a dual function in HSCs: it allows them to exit quiescence in a kinase-independent manner, whereas its kinase-dependent role in the cell cycle is beyond doubt. The kinase-dead mutant CDK6K43M regulates Egr1 in the same manner as wild-type CDK6. EGR1 is an immediate-early transcription factor with functions in stress responses, growth control, and apoptosis.40-42 Its pleiotropic functions are possible because it operates in a highly tissue-specific manner by interacting with various other transcriptional regulators.40 Mice lacking Egr1 have significantly more dividing HSCs, defining EGR1 as a central coordinator of stem cell homeostasis.13
Egr1 is frequently deregulated in transformed cells and in tumor tissue.43,44 Egr1 may function as a tumor suppressor in certain hematopoietic malignancies, including myeloid leukemia. Consistently, portions of chromosome 5 are frequently lost in myelodysplastic syndromes and AML, and Egr1 is among the genes frequently affected.45 Mice haploinsufficient for Egr1 are prone to develop myeloid disorders upon treatment with N-ethyl-N-nitrosourea.46,47 Furthermore, Egr1 is known to oppose the differentiation block inflicted by enforced c-Myc expression in myeloblastic leukemia.48 The mechanisms underlying this effect are unclear, and Egr1 has been postulated to interfere with p53.49 It has also been proposed as an upstream regulator of various tumor suppressors including TGF-β1 and phosphatase and tensin homolog.50 Whether and how the CDK6/EGR1 axis acts in other hematopoietic diseases driven by LSCs remains to be determined.
Maintaining the balance between proliferation and differentiation is not only important in recovery from hematopoietic stress, it is also a central issue in leukemia patients.22,51 HSCs play a crucial part in malignancies such as CML, which is reflected by the fact that only the transplantation of transformed HSCs is able to induce murine BCR-ABLp210+–driven myeloid leukemia. In CML, quiescent LSCs represent the leukemic stem cell compartment and are protected from conventional chemotherapy and tyrosine kinase inhibitors, allowing them to survive for many years and posing a significant challenge to therapeutic attempts.52,53 Imatinib represents a major breakthrough in therapy, although it only targets the peripheral symptoms of leukemia. The idea of inhibiting CDK6 may seem provocative, but our data show that it warrants further investigation. It may be beneficial to develop inhibitors that specifically target CDK6’s ability to regulate transcription. CDK4 and CDK6 are clearly nonredundant with regard to arousing d-HSCs and LSCs. LSCs may be more dependent on CDK6 than their nontransformed counterparts under steady-state conditions, and the difference may represent a possibility to distinguish and eliminate LSCs. Because the requirement for CDK6 is restricted to conditions of stress, including oncogenic stress, CDK6 inhibitors would not affect hematopoiesis under steady-state conditions and so might have fewer side effects than currently available forms of leukemia treatment.
The data reported in this article have been deposited in Gene Expression Omnibus (accession number GSE62012).
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sabine Fajmann and Philipp Jodl for excellent technical help, and Mathias Müller, Richard Moriggl, and all members of the SFB-F28 and SFB-F47 for helpful discussions. In particular, we thank Graham Tebb for scientific discussions and editing of the manuscript.
This work was supported by the Austrian Science Foundation (FWF) via grants to V.S. (SFB F47 and P24297-B23) and S.Z.M. (SFB F47 and P24130-B20).
Authorship
Contribution: V.S. was the principal investigator and takes primary responsibility for the article; R.S., A.H., F.B., K.K., C.S., A.T., M.P.M., M.S.R., and G.H. performed the laboratory work for this study; R.S., A.H., and V.S. wrote the manuscript; M.M., K.K., and J.Z. contributed to development of methodology; and R.S., A.H., F.B., S.Z.M., M.P.M., K.K., and M.M. were helpful in interpretation of the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for K.K. is Cambridge Institute for Medical Research and Wellcome Trust/MRC Stem Cell Institute, University of Cambridge, Hills Rd, United Kingdom.
Correspondence: Veronika Sexl, Institute of Pharmacology and Toxicology, University of Veterinary Medicine, Vienna (VetmedUni), Veterinaerplatz 1, A-1210 Vienna; e-mail: veronika.sexl@vetmeduni.ac.at.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal