Key Points
We present a practical method to ascertain CHIP that combines sequence-based and population-based filtering in the UK Biobank and All of Us.
Small changes in filtering parameters can have a large effect on the accuracy of CHIP variant classification.
Abstract
Clonal hematopoiesis of indeterminate potential (CHIP) is a common form of age-related somatic mosaicism that is associated with significant morbidity and mortality. CHIP mutations can be identified in peripheral blood samples that are sequenced using approaches that cover the whole genome, the whole exome, or targeted genetic regions; however, differentiating true CHIP mutations from sequencing artifacts and germ line variants is a considerable bioinformatic challenge. We present a stepwise method that combines filtering based on sequencing metrics, variant annotation, and population-based associations to increase the accuracy of CHIP calls. We apply this approach to ascertain CHIP in ∼550 000 individuals in the UK Biobank complete whole exome cohort and the All of Us Research Program initial whole genome release cohort. CHIP ascertainment on this scale unmasks recurrent artifactual variants and highlights the importance of specialized filtering approaches for several genes, including TET2 and ASXL1. We show how small changes in filtering parameters can considerably increase CHIP misclassification and reduce the effect size of epidemiological associations. Our high-fidelity call set refines previous population-based associations of CHIP with incident outcomes. For example, the annualized incidence of myeloid malignancy in individuals with small CHIP clones is 0.03% per year, which increases to 0.5% per year among individuals with very large CHIP clones. We also find a significantly lower prevalence of CHIP in individuals of self-reported Latino or Hispanic ethnicity in All of Us, highlighting the importance of including diverse populations. The standardization of CHIP calling will increase the fidelity of CHIP epidemiological work and is required for clinical CHIP diagnostic assays.
Introduction
Somatic mosaicism occurs across tissues as the human body ages.1 One of the best-characterized examples of this is clonal hematopoiesis of indeterminate potential (CHIP), where a somatic mutation within a hematopoietic stem cell leads to clonal production of blood cells. CHIP is defined in recent World Health Organization (WHO) and International Consensus Classification (ICC) guidelines as the presence of a somatic mutation in a myeloid neoplasm driver gene (eg, DNMT3A, TET2, ASXL1, JAK2, TP53) at a variant allele fraction (VAF) of ≥2% in an individual without a diagnosed hematologic disorder or an unexplained, persistent cytopenia.2,3 When one or more unexplained cytopenias are present, this is instead referred to as clonal cytopenia of undetermined significance (CCUS). CHIP and CCUS are premalignant lesions, and only ∼0.5% of CHIP cases progress to overt myeloid malignancy per year.4,5
Somatic CHIP variants are detected using genetic sequencing. A highly cost-effective and popular approach is to repurpose large cohort whole-genome sequencing (WGS) and whole-exome sequencing (WES) data for CHIP analyses. This method has been used to uncover the bulk of known CHIP disease associations in the population. In contrast, targeted sequencing approaches, such as gene panels, are frequently used in smaller cohort studies of CHIP and in clinical diagnostics because of their relative cost-effectiveness and greater sequencing depth, which increases accuracy.6-12 Genomes and exomes have comparatively limited sensitivity to detect small clones owing to their modest sequencing depth.13 For this reason, estimates of CHIP prevalence and effect sizes for disease associations largely depend on the sequencing method used.14
The increasing availability and decreasing cost of genetic sequencing has led to both a rapid expansion of gene sequencing for routine clinical care of patients with myeloid malignancy and an exponential growth in research studies; however, a critical gap is the assignment of pathogenicity to specific mutations. Molecular pathology interpretation of variants can be inconsistent across institutions.15,16 Published research studies have widely varying approaches to defining a sequence mutation as CHIP.4,17 In germ line genetics, publicly available large-scale reference data sets of hundreds of thousands of individuals have greatly enhanced our ability to assess pathogenicity.18 We hypothesized that a large-scale analysis of a population-scale cohort study could similarly inform the pathogenicity of variants and improve the interpretation of results for CHIP and myeloid malignancies.
Here, we provide a generalizable framework for optimizing CHIP identification across any research or clinical data set. We apply this to the 454 787-person UK Biobank (UKB) whole exome data set and to the 98 560-person All of Us whole genome data set. Although sequencing of a matched solid tissue sample is not available for large cohorts, we show linked demographic data that are available from these data sets can be leveraged to systematically identify erroneous CHIP calls and generate appropriate filtering criteria. We make these CHIP variant calls available for use by the global research community. We also provide population-scale frequency data on the mutation rate of these CHIP variants and identify recurrent sequencing artifacts.
Methods
Cohort descriptions
The UKB whole exome cohort comprised 454 787 individuals aged 40 to 70 at enrolment, when DNA was collected for sequencing.19 Participants were enrolled from 2008 to 2010 and were administered questionnaires, physical measurements, laboratory tests, and medical imaging at specified baseline and follow-up time points.20 Health outcomes since enrolment are tracked from hospitalization general practice health records and death and cancer registries. WES was performed in 2 tranches: the first 50 k using Illumina NovaSeq S2 flow cell and the second tranche of samples with the S4 flow cell to a median sequencing depth of ∼40× across sites.21 The median age at enrolment for this cohort is 58 years old (interquartile range, 50 to 63). CHIP has been ascertained by multiple groups in tranches of this data in the past using highly variable filtering criteria, resulting in differences in CHIP variant identification and prevalence estimates.22-24
The All of Us Research Program is an ongoing US-based observational cohort study.25 Linked health outcome data was pulled from participant survey questionnaires and electronic health record information. Read-level WGS data for an initial tranche of 98 560 participants was released in June 2022. WGS was performed using Illumina PCR-free whole genome technology and sequenced on the NovaSeq platform to a median sequencing depth of 40×.26 Significant emphasis was placed to ensuring All of Us WGS met clinical grade quality control specifications.27 Participants were enrolled in 2018 to 2021, and the median age at enrolment for this subset was 53 years old (interquartile range, 37-65). CHIP has not previously been ascertained in this cohort.
Putative somatic variant detection
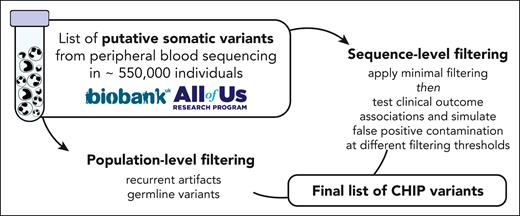
The identification of somatic variants comprises 2 major steps: putative variant identification and variant filtering (Figure 1). In the first step, a somatic variant calling pipeline is used to scan aligned sequencing files for putative somatic variants. The most commonly used somatic variant calling pipeline to detect CHIP in the research setting is Mutect2, a package within the Genome Analysis ToolKit.28 Mutect2 uses local haplotype assembly and Bayesian modeling to detect single nucleotide alterations and small indels. Mutect2 can be used for WGS, WES, or targeted sequencing data and is optimized for Illumina-based sequencing. For other sequencing platforms, a different variant caller may be necessary (eg, TorrentVariantCaller for IonTorrent data29). Other commonly used somatic variant callers include Strelka,30 VarDict,31 VarScan,32 and Shearwater.33
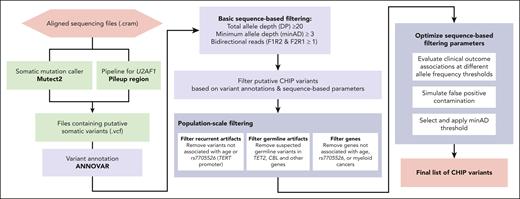
Schematic of CHIP variant ascertainment workflow. Putative somatic mutations are first identified using a somatic mutation caller and annotated for gene- and protein-level changes. Variants are then filtered based on an initial, liberal set of parameters and filtered based on gene-specific CHIP variant rules. In some genes, all loss-of-function mutations are considered putative CHIP variants, whereas in other genes, only specific missense mutations are included. Leveraging available large-scale sequencing data, we apply 3 filters to identify artifactual genes and variants. We then optimize the sequencing-based filtering parameters, yielding a final CHIP mutation call set.
Schematic of CHIP variant ascertainment workflow. Putative somatic mutations are first identified using a somatic mutation caller and annotated for gene- and protein-level changes. Variants are then filtered based on an initial, liberal set of parameters and filtered based on gene-specific CHIP variant rules. In some genes, all loss-of-function mutations are considered putative CHIP variants, whereas in other genes, only specific missense mutations are included. Leveraging available large-scale sequencing data, we apply 3 filters to identify artifactual genes and variants. We then optimize the sequencing-based filtering parameters, yielding a final CHIP mutation call set.
The foundational CHIP epidemiology papers identified a list of variants within 74 driver genes based on established variant calling from the myeloid malignancy field.34 This list specifies candidate missense and indel variants for each gene, as well as a list of genes in which truncating and splice site variants might be considered (supplemental Table 1; available on the Blood website). We limit our scan for putative CHIP variants to those contained in this list. We refer to variants in this list as the canonical CHIP driver variants, to differentiate them from more recently described gene variants observed to exhibit hematologic clonality but whose clinical consequences are less well defined.7,35,36 Of note, it is important to specify the transcript when scanning for variants, as reference transcripts are periodically updated. For example, an ETNK1 hotspot mutation that was located at N244 is now in position N155 (supplemental Table 1).
We used Mutect2 to identify putative somatic variants in 73 of the 74 canonical CHIP driver genes in the UKB and All of Us cohort aligned sequencing (CRAM) files. Altogether, 550 782 variants (1.21 variants per person) were output by Mutect2 for the UKB samples and 104 649 variants (1.06 variants per person) for All of Us samples. Variant calling pipelines such as Mutect2 cannot reliably identify variants in U2AF1 in sequencing data that are mapped to the human GRCh38 (hg38) reference genome because of an erroneous duplication of the U2AF1 locus on chromosome 21.37 A custom script was used to identify variants in U2AF1. The pileup region script counts mutated alleles present in reads that mapped to U2AF1 in one of the 2 genomic loci, and variants corresponding to prespecified hotspot locations are considered putative CHIP variants (supplemental Table 1). For samples where reads were mapped to both loci, the allelic depth was taken as the average across both sites. This yielded 65 901 U2AF1 variants in the UKB and 39 182 U2AF1 variants in All of Us, for a total of 616 683 and 143 831 putative variants, respectively. ANNOVAR was then used to annotate all putative variants for downstream filtering.38
Results
Sequencing depth-based filtering
We first apply basic filtering to remove variants with low sequencing coverage: we removed variants with a total read depth (DP) of <20, variants with a minimum read depth for the alternate allele (minAD) of <3, and variants lacking support in both forward and reverse sequencing reads. We also removed variants below the 2% VAF threshold conventionally used to define CHIP.5 This reduced the number of putative CHIP variants to 97 696 for UKB (0.21 variants per person) and 6308 for All of Us (0.06 variants per person). It may be appropriate to relax or impose further stringency of these basic filtering criteria, as we discuss later.
In large data sets, Mutect2 will output many multiallelic variants (ie, GT = 0/1/2 or 0/1/2/3). Because these are difficult to interpret and might reflect artifactual variants, many groups opt to exclude these variants from further analysis. However, some bona fide biallelic variants may appear in the multiallelic variant list; for example, more than 300 DNMT3A P904L hotspot variants and 57 PPM1D C478× hotspot variants in UKB sequencing data were misclassified as multiallelic owing to the artifactual imposition of a third allele with 0 reads at this site. Therefore, we recommend exercising caution and examining multiallelic variants separately.
Identification of false positives: sequencing artifacts
Among the list of variants that remain after basic sequence depth-based filtering, there are true CHIP variants, germ line variants, nonpathogenic somatic (passenger) variants, and sequencing artifacts. We use a multistep process to distinguish CHIP from these false positives, including filters that leverage the size of the UKB and All of Us data sets to identify recurrent false positives.
Variants that are present in ≥20 individuals in the UKB (0.004%) and in ≥15 individuals in All of Us (0.02%) were assessed for their potential to represent recurrent sequencing artifacts. For this, the association of each variant group with 2 established strong correlates of CHIP, age4,17 and a common genetic variant in the TERT promoter (rs7705526),13,24,39 was tested. Variants that were not associated with either age or rs7705526 at even a suggestive significance of P < .10 were removed from the data set as they were suspected to represent sequencing artifacts.
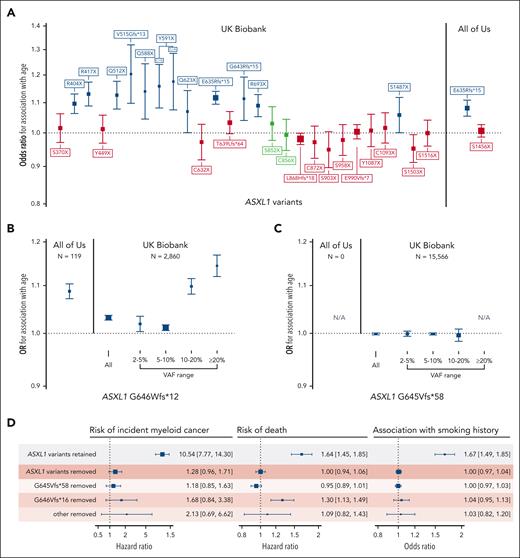
This filtering strategy proved to be particularly useful for ASXL1 variants, wherein 30 groups of truncating variants in exons 5 and 6 are reported in ≥20 people across both cohorts (Figure 2). Fifteen of the 30 variant groups examined were not associated with either age or rs7705526. We examined 2 putative variants that were present >2500 times in the UKB data set more carefully: ASXL1 p.G646Wfs∗12 and ASXL1 p.G645Vfs∗58. ASXL1 p.G646Wfs∗12 was initially thought to represent a mere sequencing artifact,40 but has been more recently confirmed to be a bona fide mutation in some cases.41,42 Montes-Moreno et al identified that sequencing methods that used PCR-only amplification introduced in vitro indels at the homopolymer locus encoding G645 and G646, whereas protocols using probe capture before PCR as well as Sanger sequencing did not.42 When these sequencing methods are compared, G646Wfs∗12 variants with a VAF ≥10% were true somatic variants, whereas all G645Vfs∗58 were artifactual. Given this, using a higher VAF threshold for ASXL1 p.G646Wfs∗12 and removing p.G645Vfs∗58 variants has been suggested as a means to remove in vitro indel contaminants.41,42 We tested different VAF thresholds for both variants in UKB and similarly found that G646Wfs∗12 variants with VAF ≥10% were associated with age (Figure 2B), whereas no G645Vfs∗58 variants VAF strata was associated with age (Figure 2C). Compared with the rest of the UKB cohort, G646Wfs∗12 variants with VAF ≥10% were associated with a 2.4-fold increased risk of death (HR, 2.4; 95% CI, 1.9-3.0) and an 18-fold increased risk of incident myeloid cancer (HR, 18.2; 95% CI, 10.7-30.9) in Cox proportional hazards models adjusted for age, age-squared, sex, smoking status, and 10 principal components of ancestry, further strengthening its credibility as a true CHIP variant. In All of Us, there were proportionally fewer G646Wfs∗12 variants (lowest VAF 9.2%), and there were no G645Vfs∗58 variants, likely because of a combination of the PCR-free sequencing library preparation methods and the longer sequencing read length. In total, there were 613 ASXL1 p.G646Wfs∗12 variants with VAF ≥10% in UKB and 119 in All of Us, making this among the top 3 most common CHIP variants in both cohorts. In many CHIP calling methods, frameshifts at homopolymer sites, such as G646Wfs∗12, are typically excluded.
Verifying the association of common putative ASXL1 variants with age can help distinguish true variants from recurrent artifacts. (A) Association of all ASXL1 variants present ≥20 times in the UKB exome data set and ≥15 times in the All of Us whole genome data set. Variants not associated with age- or a CHIP-associated TERT promoter variant (rs7705526) are colored in red. Variants associated with rs7705526 only are colored in blue. (B-C) Association of ASXL1 G646Wfs∗12 and G645Vfs∗58 with age across VAF strata identifies specific large VAF subsets of G646Wfs∗12 as somatic mutations, whereas G645Vfs∗58 appears to be an artifact of exome sequencing that is not present in All of Us. (D) There is a significant association of ASXL1 variants passing filtering with myeloid cancer, death, and smoking, but a minimal association with variants that were removed, supporting that these removed variants are artifacts.
Verifying the association of common putative ASXL1 variants with age can help distinguish true variants from recurrent artifacts. (A) Association of all ASXL1 variants present ≥20 times in the UKB exome data set and ≥15 times in the All of Us whole genome data set. Variants not associated with age- or a CHIP-associated TERT promoter variant (rs7705526) are colored in red. Variants associated with rs7705526 only are colored in blue. (B-C) Association of ASXL1 G646Wfs∗12 and G645Vfs∗58 with age across VAF strata identifies specific large VAF subsets of G646Wfs∗12 as somatic mutations, whereas G645Vfs∗58 appears to be an artifact of exome sequencing that is not present in All of Us. (D) There is a significant association of ASXL1 variants passing filtering with myeloid cancer, death, and smoking, but a minimal association with variants that were removed, supporting that these removed variants are artifacts.
In total, 152 variant groups in the UKB data (total, 38 264 variants) and 20 variant groups in the All of Us data (total, 681 variants) were not associated with either age or rs7705526. As a final step, we verified whether any of these variant groups were bona fide CHIP hotspots and identified that TP53 R175H and DNMT3A V716I were each reported in 3 or more myeloid cancer cases in COSMIC database v96. All variants not associated with age or the TERT variant, except for these 2 hotspots, were removed from both data sets and are listed in supplemental Table 2. All variants that were robustly associated with age and/or the TERT variant are indicated in supplemental Table 3.
Identification of false positives: germ line variants
Variants disrupting the catalytic domains in TET2 and CBL are considered putative CHIP variants. In traditional CHIP calling methodologies, a binomial test is used to filter out possible germ line variants in these domains; that is, a test to determine whether the measured read depth for the variant is statistically different from half of the sum of all sequencing reads at that site, as would be true for heterozygous germ line variants. We conducted a binomial test across all TET2 and CBL missense variants and flagged variants that failed the binomial test at P < .01. For many variant sites, there were multiple passing and nonpassing variants, indicating that this site might exist as an acquired CHIP variant in some and as a germ line variant in others. However, it is possible that some of the missense variants with a VAF near 50% represent large CHIP clones and not a germ line variant. In an effort to recapture some of these large VAF clones in the UKB, we examined the association of all TET2 missense variants with age and whether the addition of variants failing the binomial test improved or weakened the association with age (supplemental Figure 1). We found that the addition of large VAF clones improved the association in 3 of the 9 examined sites, TET2 H1904R, I1873T, and T1884A, suggesting that these are likely CHIP variants. These variants were exempt from the binomial test-based removal, including in All of Us.
We extended the binomial test examination to all other genes in both data sets to identify possible germ line mutations therein. For variants present more than 3 times in either data set, we examined the proportion of variants failing the binomial test. All variants in 4 variant groups, DNMT3A G298R, TP53 R110C, RUNX1 R223C and SUZ12 D725Vfs∗18, failed the binomial test at P < .01; these 159 total variants were removed from the data set. A list of variant groups where all variants failed the binomial test, that is, recurrent germ line variants, is in supplemental Table 4.
Sample quality verification
In addition to variant quality checks and rigorous filtering, it is advisable to verify the number of variants per person and inspect variant lists for samples with an unusually high variant count, which could represent a poor-quality sample. For example, in All of Us, there were 10 individuals with 4 or more mutations. In 5 of these samples, all variants appeared to be of low quality and were likely artifactual, whereas the other 5 appeared to be credible. One example of each is presented in supplemental Table 5.
Revisiting the CHIP gene list
The originally defined CHIP gene list included 74 genes43; however, larger sample sizes enable us to prune this list. We found that putative variants in 16 of the 74 genes were not positively associated with either age nor were observed in myeloid cancer cases in the either cohort and were not otherwise identified in published studies as driving myeloid CHIP (M-CHIP).34 These genes were: SF3A1, GATA1, GATA3, PTEN, SF1, STAG1, IKZF2, IKZF3, PDSS2, LUC7L2, JAK1, JAK3, GNA13, KMT2A, KMT2D, and CSF1R (supplemental Table 6). Putative variants in these genes were removed from the CHIP call set in both UKB and All of Us.
Optimizing variant level allele depth thresholds
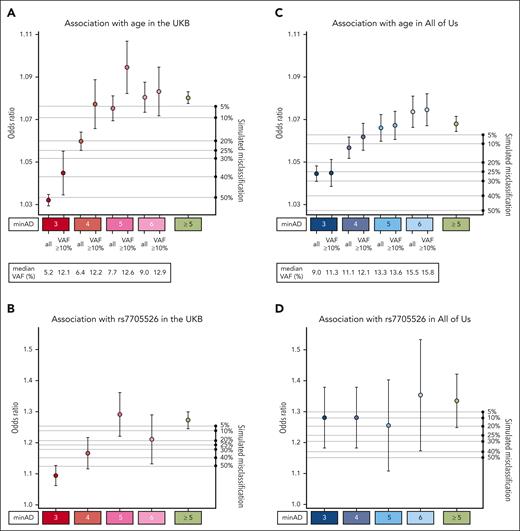
Altogether, 30 146 variants in the UKB exome cohort and 5669 variants in the All of Us first whole genome tranche passed the filtering steps. In the UKB, there were 7965 unique variants identified in this data set, 181 of which were present ≥20 times. More than half of all variants were represented fewer than 20 times in the data set (17 985 in total) and were mainly subject to the basic sequence-based filtering among the tests implemented above. In the initial filtering step, we set a relaxed minimum allele depth (minAD) threshold of 3 to increase sensitivity for CHIP variants. However, previous strategies to identify CHIP have used a minAD threshold as high as 6 to increase specificity. To identify minAD thresholds for UKB and All of Us, we first tested the associations between putative variants in minAD strata ranging from 3 to 6 with age and the TERT promoter variant (Figure 3). A minimum allele depth of 5 appeared optimal for UKB exome samples compared with lower thresholds as the associations with age and the TERT promoter variant were maximized in this stratum. To estimate how much false positive misclassification might be present in the lower strata, we ran simulations where fractions of individuals with CHIP at minAD ≥5 were randomly replaced with CHIP-free individuals (Figure 3; refer to supplemental Methods for details). The age- and TERT-variant associations for minAD 3 stratum variants were smaller than the association seen when half of the minAD ≥5 group individuals were replaced at random, suggesting that approximately 50% of the variants in the minAD 3 stratum are expected to be false positives in the UKB (Figure 3A-B). This holds true for large variants within the minAD 3 stratum (VAF ≥10%), where the predicted contamination is approximately 40%. In All of Us, the predicted contamination was lower for minAD 3: it was estimated to be between 5% and 25% based on the age and rs7705526 association analyses (Figure 3C-D). In contrast to the UKB, each minAD stratum in All of Us appeared to capture distinct VAF ranges, and the association with age gradually increases across strata.
Association of CHIP variants defined by minimum allele depth (minAD) strata with age and TERT promoter variant rs7705526. (A-D) Show the associations for strata of CHIP variants defined by minAD 3, 4, 5, and 6 with age (panels A, C) and the rs7705526 TERT promoter variant (panels B, D) in UKB and All of Us Cohorts. The right axis plots the results of a simulation experiment in which the specified proportion of samples in the minAD ≥5 CHIP data set was randomly exchanged for individuals without CHIP in the data set to estimate misclassification. Each simulation was run 20 times, and the average result is shown.
Association of CHIP variants defined by minimum allele depth (minAD) strata with age and TERT promoter variant rs7705526. (A-D) Show the associations for strata of CHIP variants defined by minAD 3, 4, 5, and 6 with age (panels A, C) and the rs7705526 TERT promoter variant (panels B, D) in UKB and All of Us Cohorts. The right axis plots the results of a simulation experiment in which the specified proportion of samples in the minAD ≥5 CHIP data set was randomly exchanged for individuals without CHIP in the data set to estimate misclassification. Each simulation was run 20 times, and the average result is shown.
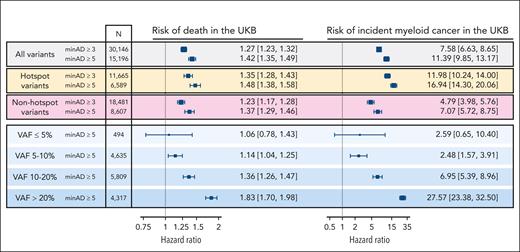
Next, we tested how minAD thresholds of 3 and 5 estimated the CHIP-associated risk of death and incident myeloid cancer risk in the UKB. A minAD threshold of 3 underestimated the risk for both outcomes compared with previous population estimates,17 including in subgroups comprising CHIP hotspot mutations (present ≥20 times in the data set) and nonhotspot mutations (present <20 times; Figure 4). As lower minAD thresholds were predicted to contain significant contamination (Figure 3) and their inclusion did not improve incident outcome predictions (supplemental Figure 2), only variants above a minAD threshold of 5 were included in our final set of UKB CHIP calls. In keeping with previous reports,4,17 CHIP was associated with an 11-fold increased risk of incident myeloid cancer (HR, 10.5; 95% CI, 9.1-12.1) and a 40% increased risk of death (HR, 1.43; 95% CI, 1.36-1.50) in the UKB. Because of limited prospective data availability in All of Us (participants were enrolled in 2018-2021), incident event analyses were not performed in All of Us. Variants above a minAD threshold of 3 were included in the final set of All of Us CHIP calls.
Associated risk of death and incident myeloid cancers with CHIP when defined using minimum sequencing allele depth (minAD) thresholds of 3 and 5 in the UKB, assessed using Cox proportional hazards regressions adjusted for age, age-squared, sex, smoking history, and 10 principal components of genetic ancestry. The risk is greater for minAD ≥5, including when hotspot variants (defined as variants observed ≥20 times in the data set) and nonhotspot variants (present <20 times) are assessed separately. The risk increases proportional to the VAF, with nearly a 10-fold increase in risk between VAF <5% and >20% strata.
Associated risk of death and incident myeloid cancers with CHIP when defined using minimum sequencing allele depth (minAD) thresholds of 3 and 5 in the UKB, assessed using Cox proportional hazards regressions adjusted for age, age-squared, sex, smoking history, and 10 principal components of genetic ancestry. The risk is greater for minAD ≥5, including when hotspot variants (defined as variants observed ≥20 times in the data set) and nonhotspot variants (present <20 times) are assessed separately. The risk increases proportional to the VAF, with nearly a 10-fold increase in risk between VAF <5% and >20% strata.
Profile of CHIP variants in the UKB and All of Us datasets
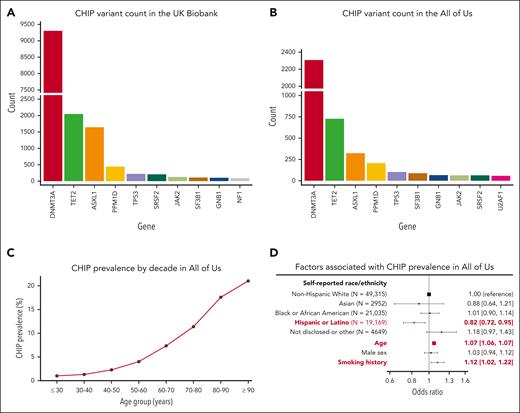
After the exclusion of individuals with hematologic malignancies before or within 6 months of study enrolment, there were 16 239 CHIP variants in 15 304 individuals in the UKB (3.6% prevalence) and 5125 CHIP variants among 4617 individuals in All of Us (4.8% prevalence). The distribution of genes affected is shown in Figure 5A-B. CHIP was detected across all decades of age, and the prevalence rose sharply with age (Figure 5C). Other risk factors previously associated with CHIP prevalence that were replicated here included smoking history23 (OR, 1.12; 95% CI, 1.02-1.22) and self-reported Hispanic or Latino ethnicity13,17 (OR, 0.82; 95% CI, 0.72-0.95; Figure 5D).
Profile of CHIP variants in the UKB and All of Us. The distribution of genes affected by CHIP in (A) the UKB and (B) All of Us are broadly similar. (C) CHIP prevalence increases with age in All of Us. (D) CHIP is associated with decreased prevalence in individuals of self-reported Hispanic or Latino ethnicity and is positively associated with age and smoking history.
Profile of CHIP variants in the UKB and All of Us. The distribution of genes affected by CHIP in (A) the UKB and (B) All of Us are broadly similar. (C) CHIP prevalence increases with age in All of Us. (D) CHIP is associated with decreased prevalence in individuals of self-reported Hispanic or Latino ethnicity and is positively associated with age and smoking history.
Finally, we explored whether certain subsets of individuals with CHIP variants may be more accurately classified as having CCUS and the prognostic implications of this. Distinguishing CHIP from CCUS is difficult in large biobanks as data is often missing to qualify a cytopenia as persistent (≥4 months) and otherwise unexplained; for example, in the UKB, more than one complete blood count (CBC) is available in <4% of participants. In single cross-sectional enrolment CBC data, CHIP was associated with higher leukocyte and platelet counts but no difference in hemoglobin levels in adjusted analyses (supplemental Figure 3A-C). Among those with CHIP, 445 (3%) had anemia without a documented cause (refer to supplemental Methods). The distribution of CHIP VAF was similar for those with unexplained anemia compared with those with explainable anemia and those without anemia, including the proportion of individuals with small CHIP clones (VAF <10%; supplemental Figure 3D). The presence of unexplained anemia is associated with a twofold to threefold increased risk of progression to myeloid cancer among those small and large CHIP clones, respectively (supplemental Figure 3E).
Discussion
CHIP and CCUS have recently been incorporated into WHO and ICC myeloid neoplasm guidelines,2,3 a step toward recognizing their importance as premalignant states with wide-ranging impacts on other organ systems. Just as the recent International Prognostic Scoring System for myelodysplastic syndromes incorporates both clinical and molecular features,44 clinicians will increasingly be tasked with the critical challenge of prognostication for patients with CHIP and CCUS. As highlighted here and by others,44,45 such prognostication will depend not only on clinical factors, such as cytopenias, or molecular features, such as clone size, but critically upon whether the mutation identified is in fact a pathogenic somatic mutation reflecting CHIP.
Here, we show how large data sets with paired genomic and demographic information can be leveraged to identify CHIP more accurately for both clinical and research applications. This strategy builds upon methodologies used in the malignant hematology field to better understand blood cancer driver mutations. We provide a set of CHIP calls for the full UKB exome data set and the All of Us interim 98k-person whole genome data set, as well as a list of recurrent false positives encountered during variant interpretation. Although our analysis was restricted to the originally described list of CHIP variants,17,43 it can be extended to other genes that have more recently been observed to exhibit clonality in blood cells.34-36
We highlight how the sequencing methodology can influence the types of artifacts detected and the filtering strategy required. For example, methods that use PCR can introduce in vitro indel artifacts at the G645 and G646 homopolymer sites in ASXL1, whereas this was not seen with the PCR-free All of Us whole genome preparation method. In both the UKB and All of Us, bona fide G646Wfs∗16 frameshifts were the most common ASXL1 variant and were strongly associated with death and myeloid cancer risk, highlighting the importance of accurately identifying this variant. We also highlight how certain hotspot variants can be missing from data sets because of known issues with reference genome assemblies (ie, U2AF1 is missing from both data sets because of hg38 error). Finally, we highlight bioinformatic strategies to identify and remove suspected germ line variants from CHIP data sets because sequencing a matched nonhematologic sample for definitive confirmation of germ line status is often not possible in large data sets.
Our work also highlights the utility of biobank scale data sets to inform ongoing efforts to define CHIP and CCUS. There is an active debate as to whether a CHIP clone in the 2% to 10% VAF range could realistically have a bearing on an unexplained cytopenia. Recognizing the limitations of available clinical data in the biobank setting, we find that cytopenias are equally frequent in individuals with small (2%-10%) CHIP clones as in individuals with large (>10%) CHIP clones; however, individuals with a cytopenia who have a large CHIP clone are at markedly increased risk for progression to malignancy. Further studies will be needed to more comprehensively elucidate factors that predict disease progression and other morbid outcomes in CHIP and CCUS.
In summary, we present a systematic approach for CHIP variant identification and validation. We anticipate that this approach and the data resource contained herein will both enhance the confidence of molecular labs and clinicians to interpret CHIP and also increase the rigor and reproducibility of research efforts to characterize CHIP at scale.
Acknowledgments
This work was supported by National Institutes of Health (NIH) Early Independence Award grant DP5 OD029586, a Burroughs Wellcome Fund Career Award for Medical Scientists, the E.P. Evans Foundation, RUNX1 Research Program, a Pew-Stewart Scholar for Cancer Research award, the Pew Charitable Trusts and the Alexander and Margaret Stewart Trust, and the Vanderbilt University Medical Center Brock Family Endowment and Young Ambassador Award, all awarded to A.J.B. C.V. received financial support from the Canadian Institutes of Health Research under the Canada Graduate Research Scholarship (RN410433–433120) and the Michael Smith Foreign Study Supplement (202106FSS-476208). A.J.S. received financial support from the NIH National Institute of Diabetes and Digestive and Kidney Diseases under the Ruth L. Kirschstein National Research Service Award F30DK127699. S.J. is supported by the Burroughs Wellcome Foundation Career Award for Medical Scientists, the Foundation Leducq, the Ludwig Center for Cancer Stem Cell Research, the Leukemia and Lymphoma Society, the American Society of Hematology Scholar Award, and the NIH Director's New Innovator Award (DP2-HL157540). M.S. receives funding from the Leukemia and Lymphoma Society, the E.P. Evans Foundation, the Biff Ruttenberg Foundation, the Adventure Alle Fund, the Beverly and George Rawlings Directorship, NIH National Cancer Institute grants 1R01CA262287 and P30 CA068485, and NIH National Institute for Occupational Safety and Health grant 1U01OH012271.
Authorship
Contribution: C.V. performed the research, analyzed data, and wrote the manuscript; A.G.B. designed the research and wrote the manuscript; T.M., J.B.H., A.N., M.M.U., J.W., B.S., A.J.S., and Y.X. assisted with performing the research and with data analysis; and M.S., C.G., M.B.L., M.J.R., B.L.E., P.N., and S.J. helped design the research and wrote the manuscript.
Conflict-of-interest disclosure: M.S. has membership on a board or advisory committee of AbbVie, Bristol Myers Squibb, CTI, Forma, Geron, Karyopharm, Novartis, Ryvu, Sierra Oncology, Taiho, Takeda, and TG Therapeutics; has patents and royalties from Boehringer Ingelheim; received research funding from ALX Oncology, Astex, Incyte, Takeda, and TG Therapeutics; owns equity in Karyopharm and Ryvu; and is a consultant in: Forma, Karyopharm, and Ryvu. B.L.E. has received research funding from Celgene, Deerfield, and Novartis and consulting fees from GRAIL; and serves on the scientific advisory boards for Skyhawk Therapeutics, Exo Therapeutics, and Neomorph Therapeutics, all unrelated to this work. S.J. is a paid consultant to Novartis, AVRO Bio, Roche Genentech, GSK, and Foresite Labs and is on the scientific advisory board to Bitterroot Bio. B.L.E., S.J., A.B., and P.N. are cofounders, equity holders, and on the scientific advisory board of TenSixteen Bio. The remaining authors declare no competing financial interests.
Correspondence: Alexander G. Bick, Division of Genetic Medicine, Vanderbilt University Medical Center, Robinson Research Bldg 550, 2200 Pierce Ave, Nashville TN; e-mail: alexander.bick@vumc.org.
References
Author notes
UKB calls and All of Us calls are in the process of being returned to the respective data sets and will be available to all registered researchers of the respective platforms upon publication. Scripts used to derive and perform the various functions in this manuscript are available at: https://github.com/briansha/Cloud_Development/tree/master/DNANexus/Mutect2 (putative somatic variant identification), https://github.com/weinstockj/pileup_region (U2AF1 putative variant identification), https://github.com/briansha/Annovar_Whitelist_Filter_WDL (annotation and filtering pipeline).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement”in accordance with 18 USC section 1734.

Comments
Re: The Case of the Missing Stopgains
We agree that the field continues to rapidly evolve, and welcome the ongoing participation in this process through collaborative contributions to the GitHub repository.
The case of the missing stopgains: nuances of sequence variant nomenclature complicate variant call filtering in Clonal Haematopoiesis of Indeterminate Potential
We read with interest the article “A practical approach to curate clonal hematopoiesis of indeterminate potential in human genetic datasets”1 by Vlasschaert, C. et al, reporting on a stepwise method of improving the accuracy of Clonal Haematopoiesis of Indeterminate Potential (CHIP) variant calls, combining filtering based on sequencing metrics, variant annotation, and population-based associations.
The pipeline consists of variant calling using Mutect22, variant annotation using ANNOVAR3 and subsequent application of a ‘whitelist’ filter using a custom Rscript. We have implemented the pipeline on a dataset of 14,000 samples. On reviewing variant calls filtered out by the pipeline, we observed an important caveat in the code provided in the ‘whitelist’ filter Rscript, when addressing loss of function mutations.
The published filtering strategy for loss of function variants is structured around the presence of STOP codons denoted by “X” or frameshifts in the ANNOVAR output.
Variants resulting in a stopgain due to nucleotide duplication introducing an immediate STOP codon without a frameshift (denoted as recommended by the Human Genome Variation Society4 simply by “*”), or stopgains due to nucleotide insertion, deletion or deletion-insertion variants without frameshifts (denoted by “del”, “ins” and “delins” respectively) are therefore erroneously filtered out by the ‘whitelist’ filter Rscript.
These include clinically relevant candidate CHIP driver variants present in our cohort e.g. the DNMT3A variant NM_175629:exon19:c.2196dupT:p.E733*, reported in 8 individuals with acute myeloid leukaemia in the COSMIC database5.
One potential solution to this problem could be to filter for annotated “stopgain” variants in the “ExonicFunc.refGene” variable of the ANNOVAR output for the relevant genes. Continual optimisation of this consensus framework with which to identify CHIP is crucial to facilitate ongoing work in this area.
1. Vlasschaert, C. et al. A practical approach to curate clonal hematopoiesis of indeterminate potential in human genetic datasets. Blood (2023) doi:10.1182/blood.2022018825.
2. Benjamin, D. et al. Calling Somatic SNVs and Indels with Mutect2. 861054 Preprint at https://doi.org/10.1101/861054 (2019).
3. Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010).
4. den Dunnen, J. T. et al. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Human Mutation 37, 564–569 (2016).
5. Tate, J. G. et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Research 47, D941–D947 (2019).