Key Points
JAK1/2 inhibitor treatment in myelofibrosis is associated with an increased risk for aggressive B-cell lymphomas.
Patients at risk have a preexisting B-cell clone in their bone marrow, which can be identified by polymerase chain reaction.
Inhibition of Janus-kinase 1/2 (JAK1/2) is a mainstay to treat myeloproliferative neoplasms (MPN). Sporadic observations reported the co-incidence of B-cell non-Hodgkin lymphomas during treatment of MPN with JAK1/2 inhibitors. We assessed 626 patients with MPN, including 69 with myelofibrosis receiving JAK1/2 inhibitors for lymphoma development. B-cell lymphomas evolved in 4 (5.8%) of 69 patients receiving JAK1/2 inhibition compared with 2 (0.36%) of 557 with conventional treatment (16-fold increased risk). A similar 15-fold increase was observed in an independent cohort of 929 patients with MPN. Considering primary myelofibrosis only (N = 216), 3 lymphomas were observed in 31 inhibitor-treated patients (9.7%) vs 1 (0.54%) of 185 control patients. Lymphomas were of aggressive B-cell type, extranodal, or leukemic with high MYC expression in the absence of JAK2 V617F or other MPN-associated mutations. Median time from initiation of inhibitor therapy to lymphoma diagnosis was 25 months. Clonal immunoglobulin gene rearrangements were already detected in the bone marrow during myelofibrosis in 16.3% of patients. Lymphomas occurring during JAK1/2 inhibitor treatment were preceded by a preexisting B-cell clone in all 3 patients tested. Sequencing verified clonal identity in 2 patients. The effects of JAK1/2 inhibition were mirrored in Stat1−/− mice: 16 of 24 mice developed a spontaneous myeloid hyperplasia with the concomitant presence of aberrant B cells. Transplantations of bone marrow from diseased mice unmasked the outgrowth of a malignant B-cell clone evolving into aggressive B-cell leukemia-lymphoma. We conclude that JAK/STAT1 pathway inhibition in myelofibrosis is associated with an elevated frequency of aggressive B-cell lymphomas. Detection of a preexisting B-cell clone may identify individuals at risk.
Introduction
Myeloproliferative neoplasms (MPNs) include polycythemia vera (PV), essential thrombocythemia, and primary myelofibrosis (PMF).1 PV and essential thrombocythemia can transform into myelofibrosis (MF). The discovery of the JAK2 V617F mutation has provided new insights into the development of MPNs and triggered the development of JAK1/2 inhibitory drugs.2,-4 Ruxolitinib, a first-in-class potent selective inhibitor of JAK1 and JAK2, improves splenomegaly, ameliorates symptoms, and is now widely used for the treatment of MF and PV.3,5 In addition to driving MPN, the JAK-STAT pathway is involved in the development of malignant lymphoma.5,-7 Recent reports point toward a slightly increased risk for lymphoid neoplasms in patients with MPN with JAK2 V617F mutations.8,,,-12 Moreover, sporadic cases of aggressive lymphomas have been reported in patients with MPN under ruxolitinib treatment.13,14 The frequency and potential causes of lymphomas under JAK2 inhibition remain unclear.
We here describe 4 cases of aggressive lymphoma including their clinical, pathological, and molecular analysis. These cases evolved under JAK1/2 inhibitor therapy between 2012 and 2016. In parallel, a mouse model was developed that recapitulates lymphoma development under MPN, using Stat1−/− mice. The transcription factor STAT1 acts downstream of JAK-kinases and is considered a tumor suppressor. In most cases, the lack of STAT1 accelerates tumor formation and is considered to counterbalance STAT3 and STAT5 activation.15,,,-19 In JAK2 V617F-driven disease, the level of activated STAT1 shapes the phenotype of the disease.7,20
Our results indicate that JAK1/2 inhibitor-associated lymphomas occur with increased frequency, have uniform clinic-pathological features, and arise from a B-cell clone that already existed during the phase of MPN. Stat1 knock-out in mice resulted in a strikingly similar course of disease, with an initial myeloproliferative disease followed by a clonal aggressive B-cell malignancy.
Methods
Patients and samples
Of 626 patients with myeloproliferative neoplasm diagnosed and treated at the Medical University of Vienna between 1997 and 2016, 69 patients with MPN have received JAK1/2 inhibitors (Ruxolitinib, Gandotinib, Fedratinib, Momelotinib) since 2009. Of these, 58 patients were included in clinical trials (ethics committee [EC] nos. 342/2009, 096/2011, 466/2011, 1032/2011, 012/2012, 0-14-12, 1910/2012, 1499/2012, 1248/2014). Data on second malignancies were retrieved from our MPN database (MUW EC 2115/2013). Clinical charts and biopsy samples of patients who developed lymphoma were carefully reviewed. Where available, material was subjected to genetic analysis. Institutional EC approval (MUW EC 553/2008) was obtained for this retrospective analysis.
MPN-associated mutations and immunoglobulin rearrangement
Peripheral blood (PB) and bone marrow (BM) samples retrieved before treatment with ruxolitinib were available from 54/69 patients. Genomic DNA was extracted using a QIAsymphony DNA Midi Kit and QIAsymphony Sp Instrument (both Qiagen). Clonality was assessed by polymerase chain reaction (PCR; as described in the BIOMED-2 study,21 using the IdentiClone IGH+ IGK B-Cell Clonality Assay Gel Detection Kit; Invivoscribe). For the detection of BCL2/IGH gene rearrangements, an IdentiClone BCL2/JH Translocation Assay Gel Detection Kit was applied according to the manufacturer's instructions. IGHV-D-J sequence was assessed by Sanger sequencing according to ERIC recommendations.22 For patients 5 and 6, clonality was assessed by PCR derived from BIOMED-2 protocols,21 followed by sequencing on MiSeq platform (Illumina). Sequences were analyzed using Vidjil software (https://app.vidjil.org).23 MPN-associated mutations were tested by the ipsogen JAK2 MutaScreen for JAK2 V617F, the ipsogen MPL W515L/K MutaScreen Kit for MPL W515L and W515K (both Qiagen), PCR fragment length analysis of exon 9 of CALR followed by Sanger sequencing for CALR mutants,24 and Sanger sequencing of exon 12 of JAK2 using a 3130xl Genomic Analyzer (Applied Biosystems). JAK2 V617F mutant allele burden was quantified by the ipsogen JAK2 MutaQuant Kit (Qiagen). In samples derived from B-cell lymphomas, DNA was isolated from paraffin-embedded lymphoma tissue. Histopathology and details on TP53 sequencing are fully described in the supplemental Appendix, available on the Blood website. Targeted next-generation sequencing of lymphoma samples (patient 1 and 2) was kindly provided by Foundation One Heme (Roche Austria GmbH, Vienna).25 The assay used DNA sequencing to interrogate 406 genes as well as selected introns of 31 genes involved in rearrangements, in addition to RNA sequencing of 265 genes.
Mice
C.Cg-Stat1tm1 (Stat1−/−)26 and C.Cg-Rag2tm1Fwa Il2rgtm1Wji (Rag2−/−γc−/−)27 mice were backcrossed on BALB/c background and maintained at the University of Veterinary Medicine, Vienna, under specific pathogen-free conditions. Experiments were approved by the institutional animal care committee and review board and conform to Austrian law (BMWFW-68.205/0218-II/3b/2012, BMWF-68.205/0103-WF/V/3b/2015, and BMWF68.205/0093-WF/V/3b/2015). Histopathology, flow cytometric analysis, transplantation studies, and other methods are fully described in the supplemental Appendix.
Cell lines, western blot, and murine b-cell clonality
Stat1−/− B-cell lines (#503, #1762, #6500, #7473) were established from diseased secondary recipients (bone marrow transplantation [BMT] of diseased primary Stat1−/− mice into Rag2−/−γc−/− animals). Stat1−/− B-cell lines were maintained analogously to BCR/ABL+ cells, as previously described.28 Lysis and western blotting was performed as described.29 Membranes were probed with antibodies for p16INK4A (56330) and β-Actin (69879; both Santa Cruz Biotechnology). Immunoglobulin gene rearrangements for clonal B-cell determination of mouse tumors were performed as described.30
Statistical analysis
Clinical data.
Categorized data are presented as absolute count and relative frequency. Continuous data are presented as mean ± standard deviation or median (25%-75% interquartile range). We compared exposed to nonexposed patients. We estimated the relative effect of exposure on the outcome by calculating a risk ratio with an exact 95% confidence interval. To test the null hypothesis of no association of the exposure with the outcome, we used the Fisher’s exact test. We assessed the influence of age and sex on the effect of JAK2 inhibitor treatment on occurrence of non-Hodgkin lymphoma (NHL), as well as on the effect of immunoglobulin rearrangement (IgR) on NHL in JAK2 inhibitor-treated patients. We estimated the univariate associations of the potential influencing variables with the exposure variable, as well as with the outcome variable. For age, we used the Student's t-test, and for sex, we used the Fisher’s exact test to test the hypothesis of no association. In a logistic regression model, we then entered each influencing variable as a covariable into the model and compared the adjusted main effect with the crude main effect.
For data management and analysis, we used MS Excel and Stata 14 (Stata Corp, College Station, TX). In general, a 2-sided P value < .05 was considered statistically significant.
Animal data.
Student’s t-test, 1-way ANOVA, Tukey’s post hoc test, and log-rank test were performed (GraphPad Prism Software). Statistical significance is indicated for each experiment specifically (*P < .05; **P < .01).
RNA-Seq
RNA-seq libraries were prepared using the SMARTer Stranded Total RNA Sample Prep Kit (Clontech, Palo Alto, CA), and 75 SE sequencing was performed on a NextSequation 500 machine (Illumina, San Diego, CA). Raw RNA-seq data (.fastq files) were adapter trimmed using TrimGalore!, followed by alignment to GRCm38 or GRCh38 using STAR algorithm.31 Quality was controlled using FastQC and RNA-seq QC tool from SeqMonk software. Differential gene expression was calculated using DESeq2.32 Cutoffs for differential gene expression were selected as false discovery rate [FDR] < 0.001 and log2fold change [FC] > 2. Murine-to-human gene symbol conversion was performed using the R package biomaRt. Heat maps were generated using ClustVis tool,33 and gene ontology enrichment analyses were performed by Ontologizer.34 Raw RNA-seq data were deposited at Gene Expression Omnibus database (GSE110219).
Results
Aggressive B-cell lymphomas during JAK1/2 inhibitor therapy in patients with MPN
Clinical characteristics and incidence of NHL in MPN cohorts are provided in Table 1.
In the Viennese cohort of 626 patients with MPN, 4 (5.8%) of 69 developed an aggressive B-cell lymphoma on JAK1/2 inhibitor treatment (patients 1-4), whereas only 2 lymphomas evolved in 557 patients (0.36%; patients 7 and 8) without inhibitor (Table 1; Table 2; supplemental Table 1). This frequency corresponds to a 16-fold increase in the probability of developing an aggressive B-cell lymphoma on JAK1/2 inhibitor therapy (95% confidence interval [CI], 3-fold to 87-fold; P = .0017). All 4 patients with lymphoma had primary (N = 3) or post-PV MF (N = 1) with a JAK2 V617F mutation (patients 1-4; Table 2) and responded clinically with symptom relief and a reduction in spleen size (not shown). Three patients (1, 2, and 4) had been pretreated with alkylating agents, and 1 patient had received ruxolitinib only. Patient 3 had also received another JAK2 inhibitor (fedratinib). The median time from start of JAK1/2 inhibitor treatment to lymphoma diagnosis was 25 months (range, 13-35 months) (Table 2).
In an independent cohort (N = 929) at Hôpital St. Louis in Paris, 2 additional patients (patients 5 and 6) developed lymphoma under ruxolitinib treatment. The frequency of lymphomas in this cohort was 0.23% in conventionally treated patients (N = 872) vs 3.51% in patients who received JAK1/2 inhibitors (N = 57), confirming the increased risk (odds ratio [OR], 15; 95% CI, 2-92; P = .0205).
Subgroup analysis of 216 patients with primary myelofibrosis in the Viennese cohort revealed that 31 patients had received JAK1/2 inhibitor therapy. Three lymphomas (9.68%) were observed under ruxolitinib, and 1 lymphoma (0.54%) developed in the 185 conventionally treated patients (OR, 19; 95% CI, 2-196; P = .01). The OR for developing lymphomas under JAK1/2 inhibitors remained unchanged after adjustment for age (OR, 21; 95% CI, 2-218) or sex (OR, 25; 95% CI, 2-266).
Distinct clinicopathological and genetic features of aggressive B-cell lymphomas
Lymphomas occurring during JAK1/2 inhibition were of aggressive CD19+ B-cell type (3 DLBCLs and 1 HGBL) and occurred at extranodal sites: Three patients (1, 2, and 3) presented with BM infiltration and PB involvement (1 only at relapse), 1 in mammary (3), and 1 in mucosal tissue (4).
Only 1 of the 4 lymphomas (patient 4) occurring under JAK1/2 inhibitor treatment stained positive for Epstein-Barr virus early RNA by in situ hybridization. None of the patients had concomitant hepatitis B or C, HIV infection, or autoimmune diseases. Details on the clinical and molecular diagnostics are provided in Table 2, with pictures of IHC stainings in supplemental Figure 1.
All 4 lymphomas showed positive MYC and p53 staining in at least 70% of the malignant cells. Patients 1 to 3 had a double-hit protein score of 2 because of additional high BCL2 expression. There was no clear association with germinal or nongerminal center cell of origin.
Fluorescence in situ hybridization detected MYC, BCL2, and/or BCL6 abnormalities in 3 patients and a 17p- deletion in 1 patient.
Material for targeted sequencing was available in 2 patients (1 and 2) and confirmed an IGH-MYC rearrangement, but revealed additional changes in CDK6, MLL, and TP53 in patient 1. In patient 2, a larger number of aberrations were found, including SOCS1-MYC and IGH-BCL2 rearrangements, as well as inactivating frameshift mutations in B2M and CDKN2A (Table 2). None of the 4 lymphomas carried a JAK2 V617F mutation even if B cells were isolated. Likewise, no other MPN-associated mutations35,-37 such as CALR, MPL, ASXL1, IDH1, IDH2, EZH2, SRSF2, SF3B1, and U2AF1 were detected in the lymphoma samples of patients 1 and 2. A variant of the ASXL1 gene (D1355E) not known to be pathogenetic was found in patient 2. No material was available for further genetic analyses in the remaining cases.
All patients had an international prognostic index38 of at least 2. Three of 4 patients responded to antilymphoma treatment, but only 1 patient is in ongoing remission, indicating a dismal prognosis. The clinical course of the 4 patients is depicted in Figure 1A-D.
Clinical courses of 4 patients who developed aggressive B-cell lymphomas during JAK1/2 inhibition. Timelines (A-D). Frequencies of immunoglobulin rearrangement in the bone marrow during myelofibrosis and lymphoma development in conventionally treated patients (N = 44) compared with patients treated with JAK1/2 inhibitor (N = 54) (E). NHL, non-Hodgkin lymphoma; CR, complete remission; CRu, complete remission unconfirmed; rNHL, remission of NHL; sAML, secondary acute myeloid leukemia.
Clinical courses of 4 patients who developed aggressive B-cell lymphomas during JAK1/2 inhibition. Timelines (A-D). Frequencies of immunoglobulin rearrangement in the bone marrow during myelofibrosis and lymphoma development in conventionally treated patients (N = 44) compared with patients treated with JAK1/2 inhibitor (N = 54) (E). NHL, non-Hodgkin lymphoma; CR, complete remission; CRu, complete remission unconfirmed; rNHL, remission of NHL; sAML, secondary acute myeloid leukemia.
The 2 lymphoma cases occurring in the absence of ruxolitinib treatment were 1 follicular lymphoma and 1 DLBCL, albeit with BM infiltration, but low MYC expression (supplemental Table 1).
Clinicopathologic characteristics were also available for 2 additional patients with lymphoma treated with JAK inhibitors at Hôpital St. Louis in Paris (patients 5 and 6 in Table 2). Both patients had aggressive DLBCL, 1 with BM infiltration. Of note, patient 6, similar to patient 4, had never received any other treatment than ruxolitinib.
Clonal B cells in the BM during the preceding MPN
We next investigated archived BM samples of 54 of 69 patients with MF before exposure to JAK1/2 inhibitors. A clonal IgR was detected by PCR during MPN in 9 (16.7%) of 54 patients (Figure 1E; supplemental Tables 2 and 3). In an age- and sex-matched control group with none or conventional therapy (N = 44), 7 patients (15.9%) tested positive for IgR (Figure 1E). This indicates the presence of clonal B cells in the bone marrow of approximately 15% of patients with PMF, regardless of treatment.
Of note, all 3 patients tested (patients 1, 2, and 4) who later developed aggressive B-cell lymphoma during treatment with ruxolitinib displayed a clonal IgR in the BM during MPN and as early as 47 to 70 months before lymphoma diagnosis (Figure 1A-D; Table 2). No material was available for patient 3.
In 2 patients (1 and 4), PCR was repeatedly positive throughout the MPN phase. In patient 1, IGH sequencing confirmed that the rearrangement of the lymphoma (IGHV4-34*01-J6*02-D3-16*01) was identical to that of the preexisting B-cell clone. In addition, an identical BCL2/IgH rearrangement (determined by sequencing) confirmed the presence of the same B-cell clone 6 years before lymphoma diagnosis in patient 2 (Figure 1B; supplemental Figure 2). Clonal IgR and BCL2/IgH rearrangement disappeared after patient 2 achieved complete response by lymphoma treatment (Figure 1B).
In contrast, only 6 (11.1%) of 54 patients who did not develop lymphoma during JAK1/2 inhibitor therapy had a positive PCR result. This relates to a 15-fold increase (95% CI, 2-fold to 128-fold) in risk of developing a B-cell lymphoma (5.9%) on JAK1/2 inhibition in patients with a detectable IgR (P = .0124). The negative predictive value of IGHV-PCR was 97%, with a sensitivity of 75% and a specificity of 88%.
In the Parisian patients with lymphoma (5 and 6), no BM was available during the MPN phase. We note that the B-cell receptor sequences of these lymphomas were also derived from IGHV4-34*01.
These data suggest that clonal IgRs are found with considerable frequency in MPN samples, and that the preexisting B-cell clone provides the basis for the evolving aggressive lymphomas under JAK1/2 inhibition.
Stat1−/− mice recapitulate the phenotype of coexisting MPN and B-cell transformation
MPN and lymphoma development was also observed in Stat1−/− mice kept under specific pathogen-free housing conditions. Stat1−/− animals show a median life span of 13.1 months compared with 28 months in wild-type (wt) mice (supplemental Figure 3A). In a cohort of 24 Stat1−/− mice, 66% developed MPN, characterized by expansion of myeloid cells and hepatosplenomegaly (Figure 2A-B). STAT1 deficiency was paralleled by the absence of Rig-I mRNA (supplemental Figure 3B), a key regulator of myelopoiesis whose loss is associated with leukemogenesis.39 In line, myeloid progenitors started to expand at the age of 4 months (supplemental Figure 3C). Concomitant with the MPN, scattered B-blasts with atypical nuclei and scant cytoplasm within a background of myeloid cells were found in lymph nodes (supplemental Figure 3D). Lowering the load of myeloid blasts by all-trans-retinoic acid led to an expected reduction of Gr1+Mac1+ cells paralleled by an increase in total B-cell numbers and enhanced number of proliferating B cells (Ki-67+) in the spleen (supplemental Figure 4A-C).
MPN and B-cell malignancy coexist in Stat1−/−mice. (A) Representative smears (BM, PB, original magnification: 40×; hematoxylin and eosin stainings) and photographs of spleens and livers. (B) Presence of Gr1+Mac1+ cells of wt and MPN+Stat1−/− animals (FACS; BM, spleen). One representative example per genotype is shown in (A) and (B). (C) FACS analysis of Gr1+Mac1+ and CD19+ (spleen). One representative example per experimental group is shown. (D) Absolute cell numbers of CD19+ cells in BMs and spleens of transplanted mice (n = 3 each). Data represent means ± SD; BM cntrl vs BM Stat1−/− recipient. **P < .01; ***P < .001). (E) Representative photographs of spleens of control (phosphate-buffered saline injected) and BMT mice.
MPN and B-cell malignancy coexist in Stat1−/−mice. (A) Representative smears (BM, PB, original magnification: 40×; hematoxylin and eosin stainings) and photographs of spleens and livers. (B) Presence of Gr1+Mac1+ cells of wt and MPN+Stat1−/− animals (FACS; BM, spleen). One representative example per genotype is shown in (A) and (B). (C) FACS analysis of Gr1+Mac1+ and CD19+ (spleen). One representative example per experimental group is shown. (D) Absolute cell numbers of CD19+ cells in BMs and spleens of transplanted mice (n = 3 each). Data represent means ± SD; BM cntrl vs BM Stat1−/− recipient. **P < .01; ***P < .001). (E) Representative photographs of spleens of control (phosphate-buffered saline injected) and BMT mice.
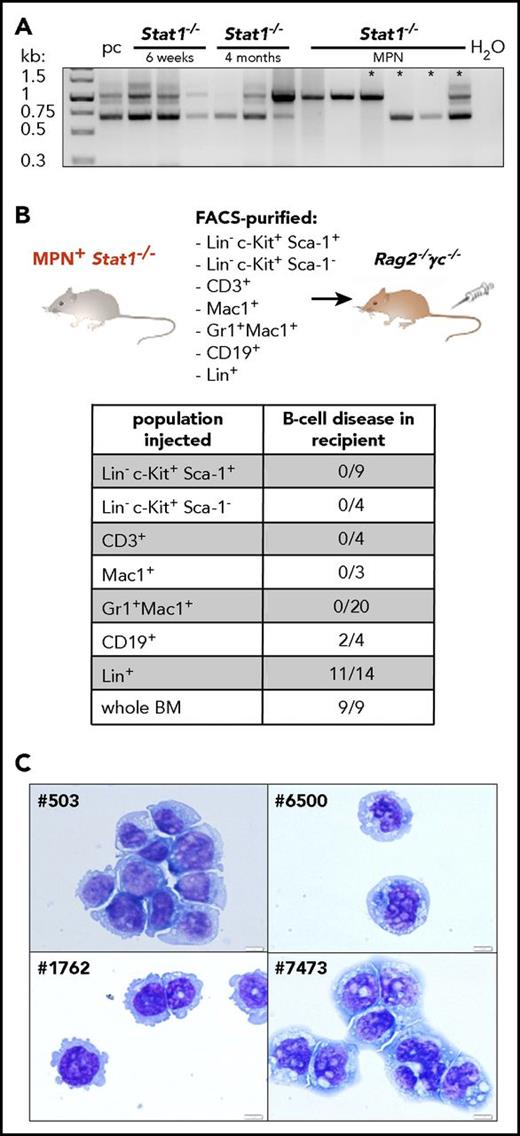
Transplantation of BM from diseased Stat1−/− mice uncovers an aggressive B-cell lymphoma
When BM cells from diseased Stat1−/− mice were injected into Rag2−/−γc−/− mice, the recipients developed pronounced hepatomegaly (supplemental Figure 5A) with significantly increased numbers of CD19+ cells (Figure 2C-D). Sizes of spleens were increased (Figure 2E) and numbers of myeloid cells in the BM and spleen reduced (supplemental Figure 5B). To investigate whether the expansion of CD19+ cells originates from a preexisting clone already present during MPN analogous to the human situation, we analyzed B-cell clonality in Stat1−/− mice at the healthy, premalignant, or MPN+ stage. Although polyclonal at early age (6 weeks, healthy), 4-month-old animals already showed a reduction of polyclonality. During MPN, 5/6 Stat1−/− mice harbored a dominant monoclonal B-cell clone (Figure 3A). In 3 of 4 mice, identical clones were detected during the phase of MPN (primary Stat1−/− mice) and in the lymphoid tissues of transplanted individuals (supplemental Figure 5C). To test whether the induction of disease roots in a leukemic cell-intrinsic malignant state, we transplanted cross-wise into Stat1−/− or wt recipients (cohorts termed “Stat1−/− into wt” and “wt into Stat1−/−”; supplemental Figure 6A). A “wt into wt” cohort was included as control. Ten (83%) of 12 mice of the “Stat1−/− into wt” cohort diseased and showed significantly elevated white blood cell counts (supplemental Figure 6B). White blood cell counts of the “wt into Stat1−/−” cohort were mildly increased, and only 1 (11.1%) of 9 mice developed disease on transplantation of wt BM. Ex vivo-derived cultivation of BM cells of the Stat1−/− into wt” cohort in a factor-free medium resulted in outgrowth of CD19+ cells (supplemental Figure 6C-D).
Stat1−/−mice recapitulate the phenotype of coexisting MPN and B-cell transformation. (A) BM of Stat1−/− mice with 6 weeks of age (n = 3), 4 months of age (n = 3), and Stat1−/− MPN+ mice (n = 6) was analyzed for D-J rearrangement of the IgH gene. Pc, polyclonal B cells (derived from splenocytes of a wt mouse). Asterisks denote samples of those mice, whose B-cell clonality was followed in subsequent transplantations (outlined in supplemental Figure 5C). (B) BMs or spleens of MPN+Stat1−/− mice were fractionated into LSK (Lin− Sca-1+c-Kit+), progenitors (Lin− c-Kit+Sca-1−), CD3+, Mac1+, Gr1+Mac1+, CD19+, and Lin+ cells and intravenous injected into Rag2−/−γc−/− mice. Table shows incidence of B-cell disease in recipient mice that had received individual populations or whole BM. (C) Cytospins of Stat1−/− B-cell lines: #503, #1762, #6500 and #7473. Original magnification: 40×. Scale bar, 10 μm.
Stat1−/−mice recapitulate the phenotype of coexisting MPN and B-cell transformation. (A) BM of Stat1−/− mice with 6 weeks of age (n = 3), 4 months of age (n = 3), and Stat1−/− MPN+ mice (n = 6) was analyzed for D-J rearrangement of the IgH gene. Pc, polyclonal B cells (derived from splenocytes of a wt mouse). Asterisks denote samples of those mice, whose B-cell clonality was followed in subsequent transplantations (outlined in supplemental Figure 5C). (B) BMs or spleens of MPN+Stat1−/− mice were fractionated into LSK (Lin− Sca-1+c-Kit+), progenitors (Lin− c-Kit+Sca-1−), CD3+, Mac1+, Gr1+Mac1+, CD19+, and Lin+ cells and intravenous injected into Rag2−/−γc−/− mice. Table shows incidence of B-cell disease in recipient mice that had received individual populations or whole BM. (C) Cytospins of Stat1−/− B-cell lines: #503, #1762, #6500 and #7473. Original magnification: 40×. Scale bar, 10 μm.
To unequivocally define CD19+ B cells as the origin of the B-cell malignancy evolving pon transplantation, we fractionated BM by fluorescence-activated cell sorting (FACS) and transplanted individual cell populations into Rag2−/−γc−/− mice (Figure 3B). Unfractionated BM was used as control and induced disease in 9 of 9 experiments. Of note, B-cell disease developed only on transplantation of CD19+ or Lin+ cells (containing B lymphocytes; Figure 3B; supplemental Figure 6E), whereas the transplantation of purified LSKs, progenitors, CD3+, Mac1+, or Gr1+Mac1+ cells failed to induce signs of malignancy. This led us to conclude that Stat1−/− mice harbor 2 individual disease entities: the initially evolving MPN that suppresses the outgrowth of a malignant B-lymphoid disease, which is unmasked on transplantation.
Aberrant Stat1−/− B cells share features with human aggressive B-cell lymphoma
The malignant B cells could be maintained in culture in vitro, were of clonal origin, and display a lymphoblast-like morphology with variable size and enlarged nuclei (Figure 3C). Stat1−/− cell lines derived from the aggressive B-cell lymphoma show reduced levels of p16INK4A and low levels of pSTAT3Y705 in the absence of pSTAT5Y694 (supplemental Figure 7A-B). FACS profiling identified the concomitant expression of B-lymphoid and stem/progenitor cell markers (IgD, SLAM, Sca-1; supplemental Figure 7C).
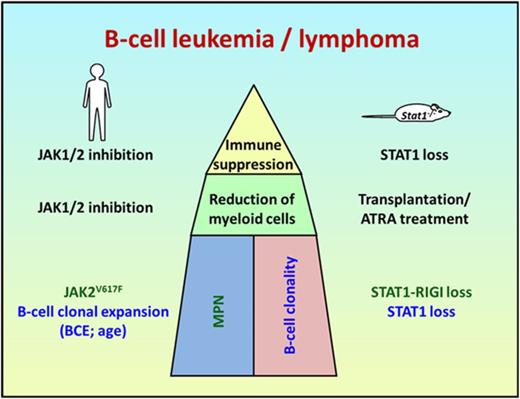
The parallels in disease development in mice and men (Figure 4A) prompted us to compare gene expression analysis by RNA-seq over both species. Control and tumor tissue from 2 patients who developed a ruxolitinib-associated B-cell lymphoma was compared with 4 murine Stat1−/− B-cell lymphoma cells and respective nontransformed controls (Figure 4B; supplemental Figure 8A). We identified 213 genes with an overlapping expression pattern (137 up- and 76 downregulated; Figure 4B). Gene ontology analysis identified deregulation of cell cycle, cell division, response to stress, or immune response as statistically significantly altered processes (Figure 4C). A complete list of differentially regulated genes is provided in supplemental Table 4. Of note, because of the high stringency (FDR<0.001 and log2FC>2), the differential expression of myc exceeded the log2FC threshold only in 1 patient (patient 1: FC, 1.87; patient 2: FC, 2.95). Thus, we decided to profile genes that are commonly mutated or deregulated in aggressive B-cell lymphomas40 in a separate analysis (supplemental Figure 8B). We identified the aberrant expression of Dsp, Prdm1, Ezh2, Smarca4, CD70, Mef2b, and Myc as commonly altered genes in mice and men.
Human aggressive B-cell lymphoma share gene expression profiles with Stat1−/−B cells. (A) Scheme summarizing and comparing disease etiology of MPN and B-cell malignancy in patients and Stat1−/− mice. (B) Identification of commonly deregulated genes in transformed murine Stat1−/− B-cell lines and in FACS-sorted CD20-positive malignant B cells from the bone marrow of patient 1 and infiltrated (>98%) lymph node from patient 2. The heat map shows 137 genes whose expression is upregulated and 76 genes whose expression is downregulated in tumor samples (TU) from patients 1 and 2 (compared with FACS-sorted and pooled B cells from the peripheral blood of 2 healthy donors or lymphocytes isolated from lymph node tissue of 1 healthy donor [Ctrl]) and in transformed murine Stat1−/− B cells (compared with FACS-sorted untransformed splenic B220+ IgM+ IgD+ CD43−Stat1−/− B cells). Colors range from blue (low expression) to red (high expression). Each row represents a single transcript, and each column represents a unique cell/tissue specimen. (C) Gene ontology enrichment analyses reveal that 213 deregulated genes are involved in cell cycle and immune response processes.
Human aggressive B-cell lymphoma share gene expression profiles with Stat1−/−B cells. (A) Scheme summarizing and comparing disease etiology of MPN and B-cell malignancy in patients and Stat1−/− mice. (B) Identification of commonly deregulated genes in transformed murine Stat1−/− B-cell lines and in FACS-sorted CD20-positive malignant B cells from the bone marrow of patient 1 and infiltrated (>98%) lymph node from patient 2. The heat map shows 137 genes whose expression is upregulated and 76 genes whose expression is downregulated in tumor samples (TU) from patients 1 and 2 (compared with FACS-sorted and pooled B cells from the peripheral blood of 2 healthy donors or lymphocytes isolated from lymph node tissue of 1 healthy donor [Ctrl]) and in transformed murine Stat1−/− B cells (compared with FACS-sorted untransformed splenic B220+ IgM+ IgD+ CD43−Stat1−/− B cells). Colors range from blue (low expression) to red (high expression). Each row represents a single transcript, and each column represents a unique cell/tissue specimen. (C) Gene ontology enrichment analyses reveal that 213 deregulated genes are involved in cell cycle and immune response processes.
Discussion
JAK1/2 inhibition is 1 of the mainstays of therapy against MF or hydroxyurea-resistant PV with considerable clinical benefit for the patients.2,-4 Here we show that JAK1/2 inhibition is associated with the risk of developing aggressive B-cell lymphoma. These lymphomas constitute a rare but consistently occurring second malignancy sharing some clinicopathological features.
Myeloproliferative neoplasms occasionally associate with lymphoproliferative disorders at diagnosis or on treatment.41 The reported prevalence of all lymphomas ranges between 0.34% and 1.6%.8,,,-12 The estimation of the occurrence of DLBCL during MPN lies between 0.12% and 0.37%.8,11,12 In line with these reports, we observed a rate of 0.36% lymphomas (1 follicular lymphoma, 1 DLBCL) in 557 Viennese patients and of 0.23% in 872 Parisian patients who were never treated with JAK1/2 inhibitors. In contrast, the frequency of lymphomas in JAK1/2 inhibitor-treated patients was significantly higher in our independent cohorts (5.8% or 3.5%), with most lymphomas developing in the PMF cohort (3 of 31; 9.7%).
Although all 4 of our patients with lymphoma were treated with ruxolitinib, 1 patient had initially been treated with another selective JAK2 inhibitor, fedratinib. It remains to be determined whether lymphoma development is restricted to ruxolitinib-treated patients or constitutes a general phenomenon on JAK1/2 inhibition. Moreover, it remains unclear whether lymphomagenesis is predominantly associated with JAK1 or JAK2 inhibition, or both.
Most patients had been pretreated with alkylating agents, a known cause of secondary malignancy including a small number of lymphoproliferative neoplasms.8,,,-12 Interestingly, patients 4 and 6 had never received any other treatment than JAK1/2 inhibition. The time from start of inhibitor treatment to lymphoma diagnosis was 13 to 35 months (median, 25 months), and is thus considered short for a second malignancy.
We note that all 4 patients in the Viennese cohort suffered from an aggressive B-cell lymphoma with extranodal involvement, including the BM and a leukemic phase in 3 patients. Expression of MYC and TP53 was high in all and BCL2 was upregulated in 3 cases. Translocations involving MYC, BCL2, and BCL6 were found. All 4 lymphomas showed similarities with 2 published cases13,14 and case 6.
The clinical features of these lymphomas are reminiscent of those occurring in immunocompromised patients except for the fact that only 1 of 4 was Epstein-Barr virus-associated. B-cell lymphomas in immunodeficient patients are aggressive, occur at extranodal sites including the BM, and have a high international prognostic index and frequent genetic alterations of MYC, BCL6, or tumor suppressors. Ruxolitinib has the ability to reduce the function of T cells and the frequency of circulating regulatory T cells and is used as an immunosuppressive agent (eg, against steroid refractory graft-versus-host disease).42,-44 It is attractive to speculate that the immunosuppressive properties of JAK1/2 inhibitors contribute to lymphoma development.42,45,,,-49
An increased prevalence of monoclonal B-cell infiltrates in untreated chronic myeloproliferative disorders has been observed previously by Pajor et al.50 In their study, 3 (13.6%) of 22 patients with MPN had a positive IgR. We confirmed these data in the bone marrow of our patients with MF. Although frequencies of IgR in patients with or without JAK1/2 inhibitor treatment were equal (16.7% and 15.9%), occurrence of lymphomas was significantly higher in the inhibitor group. Importantly, the control group was age-/sex-matched, leaving JAK1/2 inhibitor treatment as the likely trigger for the outgrowth of malignant lymphoma from a small B-cell clone. The identity of the preexisting B-cell clone was confirmed by sequencing. This opens the possibility of identifying patients at risk through PCR screening before initiation of therapy. Collection of larger data sets or incorporation of IgR into clinical trial seems warranted.50
It remains unclear whether MPN and lymphomas arise from a common ancestral clone51,52 or represent 2 independent neoplasms. In the Viennese cohort, none of the 4 lymphomas carried a JAK2 mutation, whereas the MPN had a JAK2 V617F. Furthermore, we did not detect other MPN-associated mutations53 in 2 sequenced lymphomas.
The association among JAK-STAT signaling, myeloid hyperplasia, and the occurrence of a malignant B-cell lymphoma is modeled in Stat1−/− mice. On STAT1 deficiency, myeloid hyperplasia is paralleled by the occurrence of a malignant B-cell clone, which evolves into disease on bone-marrow transplantation and gives rise to a leukemic lymphoma phenotype.
One common denominator between mice and men is immune-suppression, which is exerted in human patients by ruxolitinib. In mice, the absence of STAT1 causes dysfunctional cytotoxic T lymphocytes and NK cells. Both cell populations are required to eliminate malignant hematopoietic cells.54,-56 The evolvement of a malignant B-cell clone may thereby be facilitated. Another obvious common feature is the fact that the evolving B-cell malignancy becomes only evident on reduction of the MPN (via ruxolitinib in patients, via all-trans-retinoic acid treatment and upon transplantation in mice).
Targeted inhibition of JAK-STAT signaling appears to be required to trigger the appearance of the B-cell clone as other treatments eliminating the myeloid cell load in men do not exert a comparable effect. Although there are clear parallels for the course of disease in mice and men the underlying mechanism for the myeloproliferative disease may be different: whereas recurrent mutations (e.g.JAK2 or CALR) are involved in patients, in mice, the absence of STAT1 and its target gene Rig−I− initiates disease.57
Our study indicates that in humans and mice, the aggressive B-cell lymphomas arise from preexisting B-cell clones that are detectable at early stages of MPN. In patients, the clonal B-cell population was present as long as 6 years before overt lymphoma and preceded JAK1/2 inhibition, which offers the opportunity to determine patients at risk.
Presented in abstract form at the 23rd Congress of the European Hematology Association, Stockholm, Sweden, 15 June 2018.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Juerg Schwaller, Richard Moriggl, and Michael Freissmuth for scientific input and Graham Tebb for revision of the manuscript. For technical support, we thank Heidemarie Schuster, Michael Grubisz, Sabine Fajmann, Philipp Jodl, Gabriele Stengl, Trang Le, Martin Hilgarth, Bernadette Hilgarth, Bruno Cassinat, Veronique Meignin, and our mouse facility team. Targeted next-generation sequencing was kindly provided by Foundation One Heme, represented by Tony Fritz, Sabine Hofbauer, and Astrid Plaickner. The authors also thank the Core Facility Genomics, Medical University of Vienna, for excellent technical support.
This work was supported by the Austrian Science Fund (FWF-SFB 28 and FWF-SFB 6101, 6103, 6106, 6107 to V.S., T.D., B.S., and M.M.; FWF-SFB-47 to V.S., R.K., and P.V.; P24295-B23 to A.H.-K.; P27248-B28 to D.A.F.; and P27132-B20 to P.B.S.), the Anniversary Fund of the Austrian National Bank (OeNB; P15936 to P.B.S.), and the WWTF Precision Medicine Program LS16-034 (U.J).
Authorship
Contribution: E.P., U.J., H.G., and V.S. designed and supervised the study; S.T., A.H.-K., S.F., S.G., C.K., M.P.-M., E.M.P., A.-I.S., and K.S. performed experiments; S.T., A.H.-K., B.G., Z.B.-H., E.C.-H., E.C., G.G., G. Heller, H.H., G. Hoermann, C.K., M.-T.K., R.K., L.M., A.I.-S., C.S., W.R.S., and P.V. analyzed data; E.C.-H., T.D., D.A.F., T.K., R.K., M.M., I.S.-K., P.B.S., B.S., and V.S. provided reagents and analytic tools; J.-J.K. and E.R. provided data; A.H.-K., E.P., U.J., H.G., and V.S. designed experiments; A.H.-K., E.P., U.J., H.G., and V.S. wrote the manuscript.
Conflict-of-interest disclosure: E.P. received 2 free samples of next-generation sequencing from Foundation One Heme (Roche Austria GmbH, Vienna); U.J. received honoraria and research funding from Novartis and Roche; P.V. and H.G. received honoraria and research funding from Novartis; Z.-B.H. received honoraria from Novartis and Roche and research funding from Boehringer-Ingelheim; G.H. received research funding from BMS; G. Hoermann received honoraria and research support form Novartis; and W.R.S. received a research grant from Meda and honoraria from Celgene and Teva and participated in advisory board of Celgene, Novartis, and Jazz. The remaining authors declare no competing financial interests.
Correspondence: Ulrich Jaeger, Medical University of Vienna, Department of Medicine I, Division of Hematology and Hemostaseology, Waehringer Guertel 18-20, A-1090 Vienna, Austria; e-mail: ulrich.jaeger@meduniwien.ac.at; Veronika Sexl, University of Veterinary Medicine, Vienna, Institute of Pharmacology and Toxicology, Veterinaerplatz 1, A-1210 Vienna, Austria; e-mail: veronika.sexl@vetmeduni.ac.at; Heinz Gisslinger, Medical University of Vienna, Department of Medicine I, Division of Hematology and Hemostaseology, Waehringer Guertel 18-20, A-1090 Vienna, Austria; e-mail: heinz.gisslinger@meduniwien.ac.at.




![Figure 4. Human aggressive B-cell lymphoma share gene expression profiles with Stat1−/− B cells. (A) Scheme summarizing and comparing disease etiology of MPN and B-cell malignancy in patients and Stat1−/− mice. (B) Identification of commonly deregulated genes in transformed murine Stat1−/− B-cell lines and in FACS-sorted CD20-positive malignant B cells from the bone marrow of patient 1 and infiltrated (>98%) lymph node from patient 2. The heat map shows 137 genes whose expression is upregulated and 76 genes whose expression is downregulated in tumor samples (TU) from patients 1 and 2 (compared with FACS-sorted and pooled B cells from the peripheral blood of 2 healthy donors or lymphocytes isolated from lymph node tissue of 1 healthy donor [Ctrl]) and in transformed murine Stat1−/− B cells (compared with FACS-sorted untransformed splenic B220+ IgM+ IgD+ CD43− Stat1−/− B cells). Colors range from blue (low expression) to red (high expression). Each row represents a single transcript, and each column represents a unique cell/tissue specimen. (C) Gene ontology enrichment analyses reveal that 213 deregulated genes are involved in cell cycle and immune response processes.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/7/10.1182_blood-2017-10-810739/5/m_blood810739f4.jpeg?Expires=1768472132&Signature=C6DLzwP5yz8wGmCStIz85nYO8Zy8s7nEWLLH4axs5Of6BnnjyQINYggkc887gv51MNQQOUHhWh4ornNBtUSCJ5kW8NToBh01CIsaf3vOAgxvXlP~VptZvSNzB1mCQC~8cDEIdYweAoHstKJmBci6loxFxb1xO--Q9UhUHhwNdp-mkkZqLwAgrsvy4wzSVvV7wFkeuVmW1FD1ErakP6Pgn2OnP-f9BryIN4rcmyv6AbHE79j1qoSIgrQqIJwgqUvsu4cAJCmew-LuFMWuc2JBSSnUs81M2CiSIdlTnhNsLk4m7B8qwxk6pYkR2fkA-w2KzwputehrjwbAaj9GHdObKA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Figure 4. Human aggressive B-cell lymphoma share gene expression profiles with Stat1−/− B cells. (A) Scheme summarizing and comparing disease etiology of MPN and B-cell malignancy in patients and Stat1−/− mice. (B) Identification of commonly deregulated genes in transformed murine Stat1−/− B-cell lines and in FACS-sorted CD20-positive malignant B cells from the bone marrow of patient 1 and infiltrated (>98%) lymph node from patient 2. The heat map shows 137 genes whose expression is upregulated and 76 genes whose expression is downregulated in tumor samples (TU) from patients 1 and 2 (compared with FACS-sorted and pooled B cells from the peripheral blood of 2 healthy donors or lymphocytes isolated from lymph node tissue of 1 healthy donor [Ctrl]) and in transformed murine Stat1−/− B cells (compared with FACS-sorted untransformed splenic B220+ IgM+ IgD+ CD43− Stat1−/− B cells). Colors range from blue (low expression) to red (high expression). Each row represents a single transcript, and each column represents a unique cell/tissue specimen. (C) Gene ontology enrichment analyses reveal that 213 deregulated genes are involved in cell cycle and immune response processes.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/7/10.1182_blood-2017-10-810739/5/m_blood810739f4.jpeg?Expires=1768573549&Signature=LjR9wvUfRFvjtIOsl~7UMGsgBavw0oiMYW5RGe3E4acCkky-XxAQOvFcPhAnymifbvBKOIC7P0~hGn6tcNxpLBNgfJ3m5RYYq4A8bFHECQdj9XuYeCFeTm8iZGVZUgdXj0kX5a6aqlhMuPXJemZk-JV8qpZjC0Css6B4UXJw35ppvkMsdhOtCjreGB0pJJNxpLlZqNHVW6pnpToCzNxdDm9xbAxO5jvODfDDSXdWr352-JidRJ2jxGI7YQR2Y-UdYH2yZhNi5wRJZSCBNa7hRFPfCL9AKmQneVJ9FWALuj6EjZWNaFtN4T7xBwFhtDvHAmgWxb-tIUuQ5KCLM9Q4ag__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)