Abstract
INTRODUCTION: Mutations in TP53 can be detected in up to 16-19% patients with acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS). TP53 mutations confer adverse prognosis irrespective of currently available therapies. The clinical impact of the type, clonality and number of TP53 abnormalities is unclear. Preclinical data suggests some TP53 mutations may be associated with additional transactivation activity and increased oncogenicity.
METHODS: We evaluated 1401 patients with previously untreated AML or MDS treated at The University of Texas MD Anderson Cancer Center from 2012 to 2016. Sequencing data was obtained by use of a 28 or 53-gene targeted PCR-based next generation sequencing platform. The following mutations in TP53 where defined as GOF based on available in vitro data: R172H, R175H, R270H, G245S, R248W, G248Q and R273H. Response was defined following 2003 IWG criteria for patients with AML and 2006 revised IWG criteria for patients with MDS. Generalized linear models were used to study the association of overall response (OR), complete response (CR) and risk factors. Kaplan-Meier produce limit method was used to estimate the median overall survival (OS).
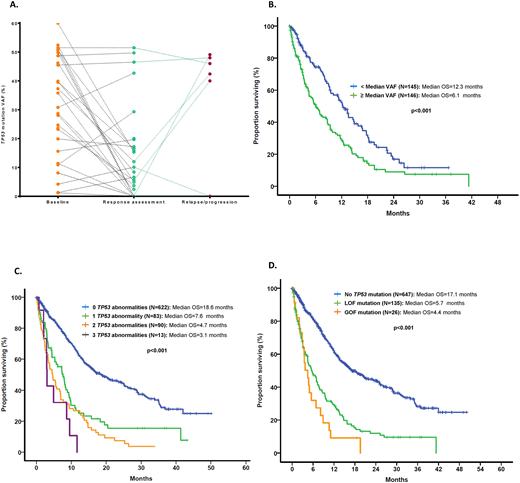
RESULTS: A total of 593 (42%) patients had MDS and 808 (56%) had AML. In a total of 984 (70%) patients, data on therapy with sufficient follow up and response evaluation was available, with 494 (35%) patients receiving therapy with hypomethylating agents (HMAs) and 373 (27%) with chemotherapy regimens. Among evaluated MDS pts, based on the Revised-IPSS prognostic model, 62 (11%) had very-low risk, 149 (25%) had low, 121 (20%) intermediate, 109 (18%) high and 152 (26%) very-high risk. A total of 167 (28%) pts with MDS and 223 (26%) pts with AML had complex karyotype. A total of 405 mutations in TP53 were detected among 151 (26%) and 161 (20%) patients with MDS and AML, respectively. Mutations included 332 (82%) missense, 26 (6%) nonsense, 34 (8%) frameshift insertions or deletions and 13 (3%) splice-site mutations. The most prevalent mutation was R273H (MDS: n=10, AML: n=9) followed by R248W (MDS: n=8, AML: n=8), Y220C (MDS: n=7, AML: n=8) and R175H (MDS: n=7, AML: n=6). Median variant allele frequency (VAF) of TP53 mutations was 39% (range 1-94). Among patients with TP53-mutant disease: 1 TP53 mutation was identified in 105 (70%) and 126 (78%) MDS and AML pts respectively, 2 in 44 (29%) and 34 (21%) and 3 in 2 (1%) and 1 (1%) respectively. The median difference in VAF among detectable mutations in pts with 2 TP53 mutations was 3.9% [95% CI 4.9-14%]. Among double TP53-mutant pts, known GOF mutations where always found in association with a LOF mutation with no pts having 2 detectable GOF mutations. Additionally, 66 (11%) MDS and 105 (13%) AML patients had TP53 deletions evidenced by presence of monosomy 17 or del(17p). Presence of a TP53 mutation was associated with loss of TP53 locus by cytogenetic abnormality (r=0.492, p<0.001). However, patients with multiple detectable TP53 mutations were less likely to have co-occurring chr17 abnormalities (79% vs 21%, OR 0.48, CI 0.29-0.81, p=0.01). Sequential bone marrow sequencing through the course of disease evolution was available in 75 pts. Dynamics of TP53 VAF at time or response and subsequent relapse in evaluable pts is shown in Figure A. Median follow up was 10.4 months (range 0-167 months) for MDS pts and 7.8 months (range 0-50) in AML pts. Presence of TP53 mutations with high VAF (defined as VAF > median VAF) was associated with significantly worse median OS (Figure B). Presence of more than 1 TP53 abnormality was not associated with worse OS, LFS or PFS in pts with MDS but predicted for shorter OS in pts with AML (Figure C). In addition, GOF were associated with worse OS than LOF mutations in pts with AML (Figure D), but not in MDS. Presence and number of TP53 mutations did not predict for response to HMAs. In addition, clearance of mutation at the time of response was associated with improved OS (HR 0.26, 95% CI 0.08-0.78, p=0.016).
CONCLUSION: Presence of multiple TP53 abnormalities can be observed in up to 13% of patients with MDS or AML. The number of TP53 abnormalities does not seem to affect the survival of patients with MDS, but is associated with worse OS in AML. In addition, although GOF mutations do not seem to affect outcome of pts with MDS, these mutations were associated with worse OS than LOF in pts with AML. Clonal size of TP53 mutations as well as their clearance in therapy correlate with survival outcomes.
Sasaki:Otsuka Pharmaceutical: Honoraria. Kadia:Jazz: Consultancy, Research Funding; Jazz: Consultancy, Research Funding; Amgen: Consultancy, Research Funding; Celgene: Research Funding; Amgen: Consultancy, Research Funding; Novartis: Consultancy; Abbvie: Consultancy; Pfizer: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; Abbvie: Consultancy; Takeda: Consultancy; Novartis: Consultancy; BMS: Research Funding; BMS: Research Funding; Celgene: Research Funding; Takeda: Consultancy. Ravandi:Astellas Pharmaceuticals: Consultancy, Honoraria; Abbvie: Research Funding; Jazz: Honoraria; Abbvie: Research Funding; Bristol-Myers Squibb: Research Funding; Jazz: Honoraria; Macrogenix: Honoraria, Research Funding; Sunesis: Honoraria; Sunesis: Honoraria; Orsenix: Honoraria; Amgen: Honoraria, Research Funding, Speakers Bureau; Seattle Genetics: Research Funding; Amgen: Honoraria, Research Funding, Speakers Bureau; Seattle Genetics: Research Funding; Macrogenix: Honoraria, Research Funding; Bristol-Myers Squibb: Research Funding; Astellas Pharmaceuticals: Consultancy, Honoraria; Xencor: Research Funding; Orsenix: Honoraria; Xencor: Research Funding. Cortes:novartis: Research Funding. Daver:Novartis: Research Funding; Incyte: Consultancy; Karyopharm: Research Funding; Pfizer: Consultancy; ImmunoGen: Consultancy; Daiichi-Sankyo: Research Funding; Otsuka: Consultancy; BMS: Research Funding; Sunesis: Research Funding; Sunesis: Consultancy; Alexion: Consultancy; Novartis: Consultancy; Incyte: Research Funding; Kiromic: Research Funding; ARIAD: Research Funding; Karyopharm: Consultancy; Pfizer: Research Funding. DiNardo:Karyopharm: Honoraria; Medimmune: Honoraria; Bayer: Honoraria; Celgene: Honoraria; Agios: Consultancy; Abbvie: Honoraria. Jabbour:novartis: Research Funding. Pemmaraju:daiichi sankyo: Research Funding; samus: Research Funding; cellectis: Research Funding; abbvie: Research Funding; stemline: Consultancy, Honoraria, Research Funding; Affymetrix: Research Funding; novartis: Research Funding; plexxikon: Research Funding; celgene: Consultancy, Honoraria; SagerStrong Foundation: Research Funding. Konopleva:cellectis: Research Funding; Immunogen: Research Funding; Stemline Therapeutics: Research Funding; abbvie: Research Funding. Colla:Abbvie: Research Funding. Andreeff:Eutropics: Equity Ownership, Membership on an entity's Board of Directors or advisory committees; Jazz Pharma: Consultancy; Aptose: Equity Ownership, Membership on an entity's Board of Directors or advisory committees; Daiichi-Sankyo: Consultancy, Patents & Royalties: MDM2 inhibitor activity patent, Research Funding; United Therapeutics: Patents & Royalties: GD2 inhibition in breast cancer ; Reata: Equity Ownership; Celgene: Consultancy; Oncolyze: Equity Ownership; Oncoceutics: Equity Ownership, Membership on an entity's Board of Directors or advisory committees; Astra Zeneca: Research Funding; Amgen: Consultancy, Research Funding; SentiBio: Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal